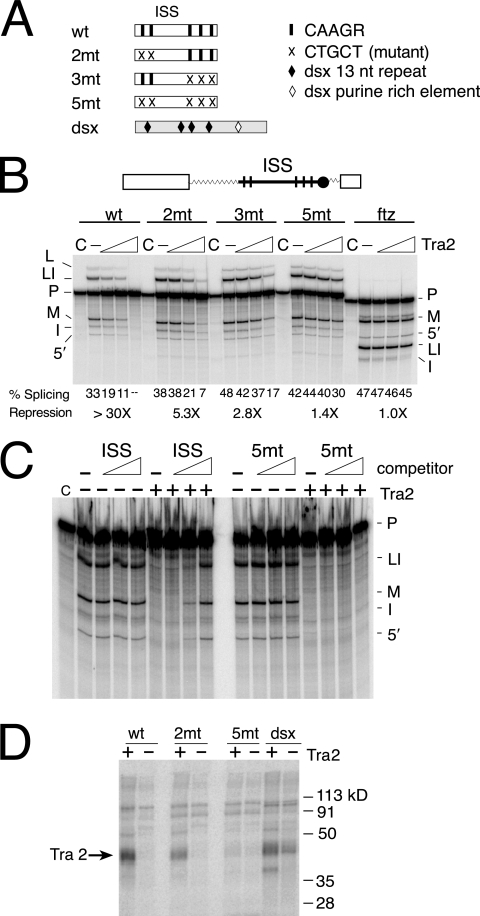
FIG. 3.
The CAAGR repeats in the ISS are required for Tra2 repression and binding. (A) A schematic showing the positions of point mutations affecting repeats in the ISS is shown. The black bars indicate CAAGR repeats, and the crosses indicate mutated repeats. Sequences from the dsx splicing enhancer were used as a positive control in UV cross-linking assays. (B) Splicing assays on substrates with ISS mutations are shown. Above the gel is a diagram of ftz-ISS, the base substrate used in all assays. Within this substrate, different forms of ISS including wt, 2mt, 3mt, and 5mt are tested for response to recombinant Tra2 protein. The ftz RNA substrate (ftz) served as a negative control. Splicing percentages and maximal relative (n-fold) repression are listed below the gel. Products and intermediates are labeled as described for Fig. 1. (C) Unlabeled ISS or 5mt RNA fragments (120 to 197 nt) were used as competitors in reactions containing repressive levels of Tra2 protein and the radiolabeled substrate ftz-M1 1-208. Increasing height of the triangle indicates increasing amount of RNA competitors. Note that splicing is restored by the wild-type ISS but not by the 5mt RNA with altered repeat elements. Reactions without Tra2 are shown as controls for nonspecific repression of splicing. (D) Labeled wt, 2mt, and 5mt ISS RNA fragments (120 to 197 nt) were UV cross-linked in Drosophila S2 nuclear extract under splicing conditions both with (+) and without (−) recombinant Tra2 protein present. Cross-linking reactions carried out on RNA from the dsx splicing enhancer are shown for comparison. The position of cross-linked recombinant Tra2 protein is indicated by the arrow.