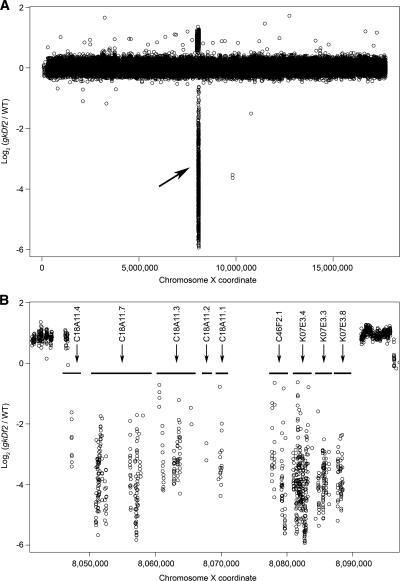
Figure 1.
Detection of a 50-kb homozygous viable deletion in gkDf2. (A) Normalized log2 ratios (gkDf2/WT) of the average fluorescent intensities for each of the 92,209 forward and reverse pairs of probes to the X chromosome are represented by circles. The deletion is identified by negative log2 ratios and indicated by an arrow. (B) A higher resolution view of fluorescence ratios for probe pairs targeting the 50-kb deletion. Horizontal bars indicate the positions of the nine genes targeted by the deletion. Duplications of sequences flanking the deletion are indicated by positive log2 ratios. Adjacent 50-mer probes in this region overlap by as much as 44 bp.