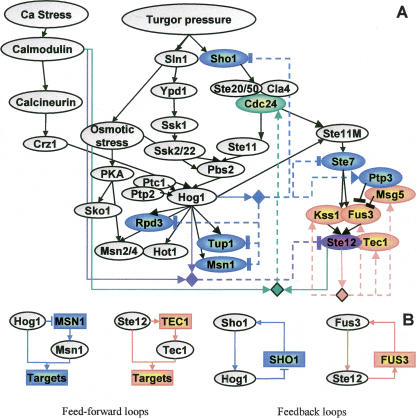
Figure 7.
Complex transcriptional feedback in the yeast osmotic network model. (A) We highlight in color model variables whose corresponding genes were included in a module. Regulatory units are shown as diamonds, where the incoming arcs indicate the regulators they contain, and outcoming dashed arcs indicate their (direct or indirect) targets. For each regulatory unit, we use a different color for its target genes and the relevant edges. For example, the unit of Ste12 (orange) has TEC1, FUS3, KSS1, and MSG5 among its targets. Unlike previous maps, the same diamond might represent several different regulatory logics, and the arcs distinguish between positive (→) and negative (⫞) feedback. The rich circuitry observed is probably part of the cellular adaptation and provides rapid and transient response to osmotic stress. (B) A few network motifs discovered in A. Rectangles indicate target genes, and ovals are proteins.