Abstract
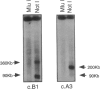
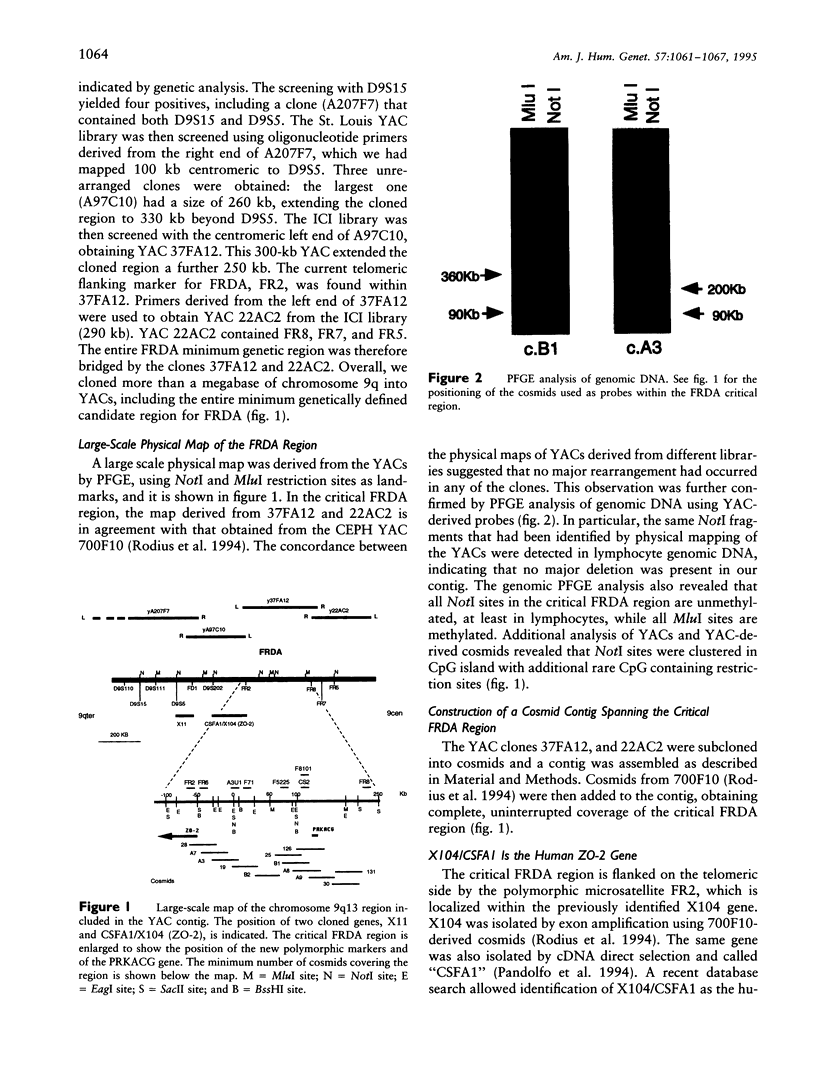
By analysis of crossovers in key recombinant families and by homozygosity analysis of inbred families, the Friedreich ataxia (FRDA) locus was localized in a 300-kb interval between the X104 gene and the microsatellite marker FR8 (D9S888). By homology searches of the sequence databases, we identified X104 as the human tight junction protein ZO-2 gene. We generated a largescale physical map of the FRDA region by pulsed-field gel electrophoresis analysis of genomic DNA and of three YAC clones derived from different libraries, and we constructed an uninterrupted cosmid contig spanning the FRDA locus. The cAMP-dependent protein kinase γ-catalytic subunit gene was identified within the critical FRDA interval, but it was excluded as candidate because of its biological properties and because of lack of mutations in FRDA patients. Six new polymorphic markers were isolated between FR2 (D9S886) and FR8 (D9S888), which were used for homozygosity analysis in a family in which parents of an affected child are distantly related. An ancient recombination involving the centromeric FRDA flanking markers had been previously demonstrated in this family. Homozygosity analysis indicated that the FRDA gene is localized in the telomeric 150 kb of the FR2-FR8 interval.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Chamberlain S., Farrall M., Shaw J., Wilkes D., Carvajal J., Hillerman R., Doudney K., Harding A. E., Williamson R., Sirugo G. Genetic recombination events which position the Friedreich ataxia locus proximal to the D9S15/D9S5 linkage group on chromosome 9q. Am J Hum Genet. 1993 Jan;52(1):99–109. [PMC free article] [PubMed] [Google Scholar]
- Chamberlain S., Shaw J., Rowland A., Wallis J., South S., Nakamura Y., von Gabain A., Farrall M., Williamson R. Mapping of mutation causing Friedreich's ataxia to human chromosome 9. Nature. 1988 Jul 21;334(6179):248–250. doi: 10.1038/334248a0. [DOI] [PubMed] [Google Scholar]
- Duclos F., Rodius F., Wrogemann K., Mandel J. L., Koenig M. The Friedreich ataxia region: characterization of two novel genes and reduction of the critical region to 300 kb. Hum Mol Genet. 1994 Jun;3(6):909–914. doi: 10.1093/hmg/3.6.909. [DOI] [PubMed] [Google Scholar]
- Foss K. B., Simard J., Bérubé D., Beebe S. J., Sandberg M., Grzeschik K. H., Gagné R., Hansson V., Jahnsen T. Localization of the catalytic subunit C gamma of the cAMP-dependent protein kinase gene (PRKACG) to human chromosome region 9q13. Cytogenet Cell Genet. 1992;60(1):22–25. doi: 10.1159/000133286. [DOI] [PubMed] [Google Scholar]
- Fujita R., Agid Y., Trouillas P., Seck A., Tommasi-Davenas C., Driesel A. J., Olek K., Grzeschik K. H., Nakamura Y., Mandel J. L. Confirmation of linkage of Friedreich ataxia to chromosome 9 and identification of a new closely linked marker. Genomics. 1989 Jan;4(1):110–111. doi: 10.1016/0888-7543(89)90323-6. [DOI] [PubMed] [Google Scholar]
- Fujita R., Sirugo G., Duclos F., Abderrahim H., Le Paslier D., Cohen D., Brownstein B. H., Schlessinger D., Mandel J. L., Koenig M. A 530kb YAC contig tightly linked to the Friedreich ataxia locus contains five CpG clusters and a new highly polymorphic microsatellite. Hum Genet. 1992 Jul;89(5):531–538. doi: 10.1007/BF00219179. [DOI] [PubMed] [Google Scholar]
- Geoffroy G., Barbeau A., Breton G., Lemieux B., Aube M., Leger C., Bouchard J. P. Clinical description and roentgenologic evaluation of patients with Friedreich's ataxia. Can J Neurol Sci. 1976 Nov;3(4):279–286. doi: 10.1017/s0317167100025464. [DOI] [PubMed] [Google Scholar]
- Harding A. E. Friedreich's ataxia: a clinical and genetic study of 90 families with an analysis of early diagnostic criteria and intrafamilial clustering of clinical features. Brain. 1981 Sep;104(3):589–620. doi: 10.1093/brain/104.3.589. [DOI] [PubMed] [Google Scholar]
- Jesaitis L. A., Goodenough D. A. Molecular characterization and tissue distribution of ZO-2, a tight junction protein homologous to ZO-1 and the Drosophila discs-large tumor suppressor protein. J Cell Biol. 1994 Mar;124(6):949–961. doi: 10.1083/jcb.124.6.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leone M., Rocca W. A., Rosso M. G., Mantel N., Schoenberg B. S., Schiffer D. Friedreich's disease: survival analysis in an Italian population. Neurology. 1988 Sep;38(9):1433–1438. doi: 10.1212/wnl.38.9.1433. [DOI] [PubMed] [Google Scholar]
- Monrós E., Smeyers P., Rodius F., Cañizares J., Moltó M. D., Vilchez J. J., Pandolfo M., Lopez-Arlandis J., de Frutos R., Prieto F. Mapping of Friedreich's ataxia locus by identification of recombination events in patients homozygous by descent. Eur J Hum Genet. 1994;2(4):291–299. doi: 10.1159/000472373. [DOI] [PubMed] [Google Scholar]
- Pandolfo M., Pizzuti A., Redolfi E., Munaro M., Di Donato S., Cavalcanti F., Filla A., Monticelli A., Pianese L., Cocozza S. Isolation of a new gene in the Friedreich ataxia candidate region on human chromosome 9 by cDNA direct selection. Biochem Med Metab Biol. 1994 Aug;52(2):115–119. doi: 10.1006/bmmb.1994.1041. [DOI] [PubMed] [Google Scholar]
- Pandolfo M., Sirugo G., Antonelli A., Weitnauer L., Ferretti L., Leone M., Dones I., Cerino A., Fujita R., Hanauer A. Friedreich ataxia in Italian families: genetic homogeneity and linkage disequilibrium with the marker loci D9S5 and D9S15. Am J Hum Genet. 1990 Aug;47(2):228–235. [PMC free article] [PubMed] [Google Scholar]
- Rodius F., Duclos F., Wrogemann K., Le Paslier D., Ougen P., Billault A., Belal S., Musenger C., Brice A., Dürr A. Recombinations in individuals homozygous by descent localize the Friedreich ataxia locus in a cloned 450-kb interval. Am J Hum Genet. 1994 Jun;54(6):1050–1059. [PMC free article] [PubMed] [Google Scholar]
- Sirugo G., Cocozza S., Brice A., Cavalcanti F., De Michele G., Dones I., Filla A., Koenig M., Lorenzetti D., Monticelli A. Linkage disequilibrium analysis of Friedreich's ataxia in 140 Caucasian families: positioning of the disease locus and evaluation of allelic heterogeneity. Eur J Hum Genet. 1993;1(2):133–143. doi: 10.1159/000472400. [DOI] [PubMed] [Google Scholar]
- Sirugo G., Keats B., Fujita R., Duclos F., Purohit K., Koenig M., Mandel J. L. Friedreich ataxia in Louisiana Acadians: demonstration of a founder effect by analysis of microsatellite-generated extended haplotypes. Am J Hum Genet. 1992 Mar;50(3):559–566. [PMC free article] [PubMed] [Google Scholar]