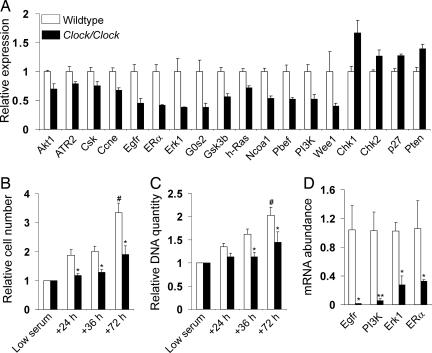
Fig. 4.
DNA synthesis and cell proliferation is diminished in Clock primary cells. (A) The average expression intensities from 24-h microarray data are shown for many genes that are involved in cell proliferation and exhibit significantly (P < 0.05; see SI Table 8) changed expression in Clock liver. (B and C) MEFs from WT and Clock embryos were cultured in 2% BCS for 48 h and then switched to media containing 10% FBS. The number of live cells (B) and quantity of DNA (C) were measured during low-serum and high-serum (+24 h, +36 h, +72 h) conditions. At least three wells per culture were measured at each time point, and all data were normalized to the average low-serum values for each culture. Asterisks represent the significant difference observed for both the relative cell number and relative DNA quantity of WT MEF cells versus Clock MEF cells at +36 h and +72 h, and the relative cell number of WT MEFs versus Clock MEFs at +24 h (∗, effect of genotype; two-way ANOVA). In addition, WT MEFs exhibited significantly higher relative cell number and DNA quantitation at +72h, compared with other time points (#, effect of time; two-way ANOVA). Error bars represent standard deviation from the means of normalized data from 12 independent WT and 10 independent Clock MEF cultures. (D) mRNA expression from MEFs grown in 10% FBS was determined by RT-PCR. Expression of the proproliferative genes Erk1, ERα, Egfr, and PI3K was significantly reduced in Clock MEFs (∗, P < 0.05; ∗∗,P < 0.01; Student's t test).