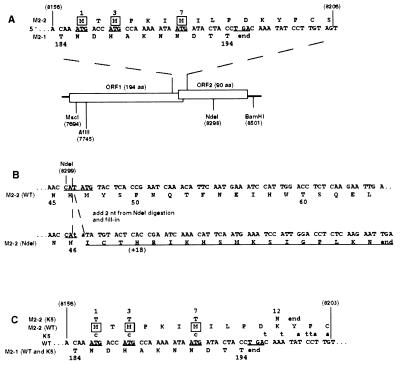
Figure 1.
Construction of the NdeI and K5 mutations, which interrupt M2 ORF2. Nucleotide sequences are in positive sense and blocked in triplets according to amino acid coding in ORF2. Nucleotide positions relative to the complete 15,223-nt recombinant antigenome are in parentheses; the other numbers refer to amino acid positions in the 194-aa M2–1 protein or 90-aa M2–2 protein. (A) Diagram of the two overlapping M2 ORFs. In the sequence at the top, the three potential translational start sites for M2–2 are underlined, and their encoded methionine residues are boxed. The termination codon for ORF1 is also underlined. In the diagram, restriction sites used for mutagenesis and cloning are indicated. (B) Construction of the NdeI mutation. The NdeI site at position 8299 in the middle of the M2–2 ORF was opened, filled in, and religated, which added 2 nt (lower case) to codon 47 of M2–2. This shifted the register to another reading frame, which was open for 18 additional codons encoding non-M2–2 amino acids (underlined). (C) Construction of the K5 mutation. The sequence shows the junction between ORF1 and ORF2, as in A. Potential ORF2 initiation codons in the wt parent are underlined, as is the ORF1 termination codon. Nucleotide changes in K5 are indicated above their wt counterparts. The three potential initiation codons for ORF2, codons 1, 3, and 7, were changed to ACG, which had no effect on amino acid coding in ORF1. The next potential methionyl start site in ORF2 is at codon 30. In addition, stop codons were introduced into all three frames immediately downstream of the M2–1 termination codon. In combination, these mutations had the effect of changing M2–2 amino acid 12 from K to N and terminating at codon 13.