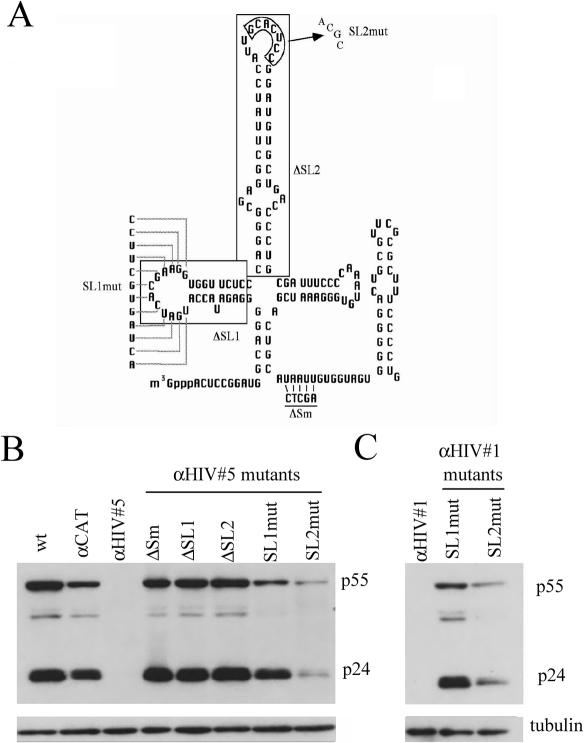
Figure 3.
HIV-1 Suppression is dependent upon the integrity of the U1 snRNP. (A) Schematic of U1 snRNA structure with the location and nature of U1 mutants indicated. Shown are the regions of α-HIV U1 #1 and #5 that were mutated to identify the protein components of the snRNP required to suppress viral gene expression. Mutations were as follows: deletion of stem–loop 1 (ΔSL1), deletion of stem–loop 2 (ΔSL2), substitutions in stem–loop 1 (SL1mut), substitutions in the loop region of stem–loop 2 (SL2mut) and mutations in the Sm binding region that abolishes assembly of the Sm core complex (ΔSm). (B and C) Effect of anti-HIV-1 #5/#1 mutation on Suppression of HIV-1 Gene Expression Cells were transfected with a plasmid containing HIV-1 proviral DNA (HIV-1 Hxb2 R-/RI-) and mutants of anti-HIV #5 (B) or #1 (C). Forty-eight hours post-transfection, cells were harvested and level of p24 production assessed by western blot.