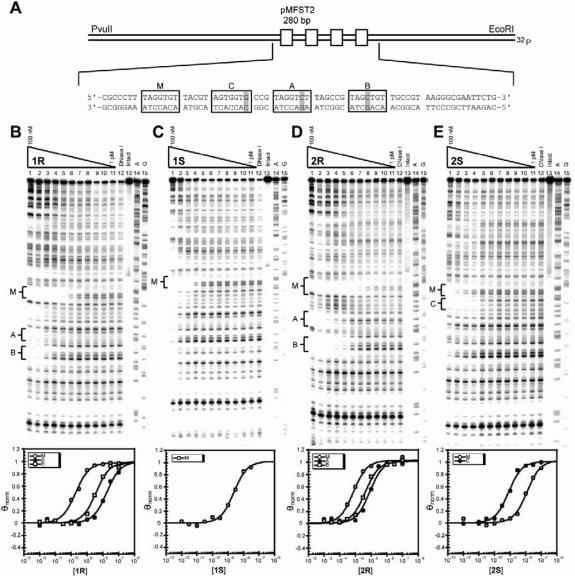
Figure 2.
Plasmid design and DNA-binding properties of the parent polyamides. (A) Illustration of the designed portion of the EcoRI/PvuII restriction fragment derived from plasmid pMFST2. The four polyamide binding sites are indicated by boxes. Single base pair mismatches are indicated by shaded regions. Site C is a mismatch site for a polyamide binding in reverse. (B–E) Quantitative DNase I footprint titration experiments for polyamides 1R (B), 1S (C), 2R (D), 2S (E) on the 280 bp, 5′ end-labeled PCR product of plasmid pMFST2: lanes 1–11, 100, 30, 10, 3, 1nM; 300, 100, 30, 10, 3 and 1 (M−1) polyamide, respectively; lane 12, DNase I standard; lane 13, intact DNA; lane 14, A reaction; lane 15, G reaction. Each footprinting gel is accompanied by its respective binding isotherms for the indicated sites (below).