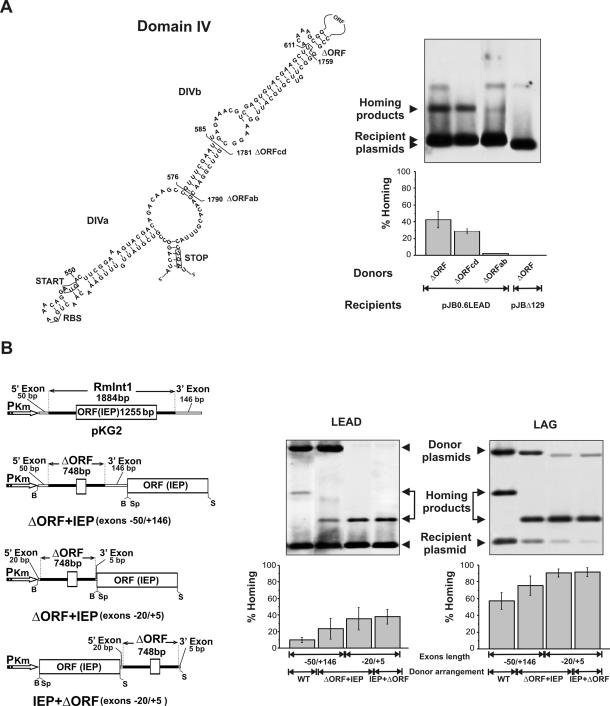
Figure 2.
Homing efficiency of wild-type RmInt1 and ΔORF-derivatives. (A) Deletion analysis of RmInt1 DIV. The predicted secondary structure of intron DIV is shown with indication of subdomains DIVa and DIVb. Start and stop codons of the IEP ORF are boxed and brackets demarcate the internal deletions (ΔORF, ΔORFcd and ΔORFab) cloned downstream of the IEP coding sequence in the donor constructs. For homing assays plasmid pools from RMO17 cells harbouring donor and recipient (pJB0.6LEAD) plasmids were analyzed by Southern hybridization with a DNA probe specific to the insertion sequence ISRm2011-2. Recipient plasmid pJBΔ129 was used as negative control in the assays. Target invasion rates in each homing assay were calculated as described in Materials and Methods and plotted in the graph-bar shown below the blot. (B) Effect of different donor genetic arrangements on intron homing. The configuration of the donor constructs is diagrammed to the left. PKm, promoter of Km resistance gene; B, BamHI; Sp, SpeI; S, SacI. Homing efficiency on recipient plasmids pJB0.6LEAD and pJB0.6LAG was quantified as described in A. The Southern blots are shown to the right along with the corresponding histograms. Features of the donor constructs are indicated bellow the plots.The hybridization signal corresponding to the donor plasmid with short flanking exons is variable and depends on the specific activity of the probe.