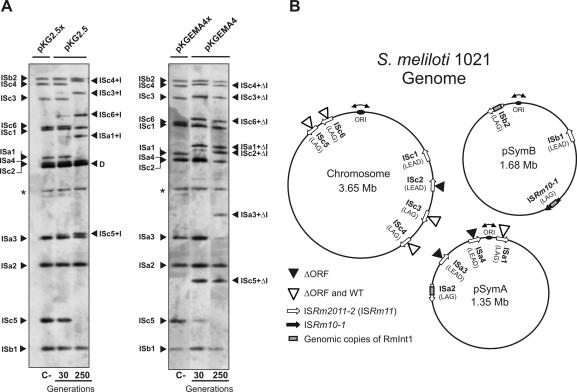
Figure 4.
Retrohoming of RmInt1 into the genome of the S.meliloti reference strain 1021. (A) Southern hybridizations of BamHI-digested DNA from 1021 transconjugants harboring plasmids pKG2.5 or pKGEMA4 grown for 30 and 250 generations. Membranes were probed with a DNA fragment specific to the 3′ end of ISRm2011-2. First lane of each panel (C-) shows an ISRm2011-2 (IS) fingerprint of 1021 derivatives expressing non-functional introns from plasmids pKG2.5x or pKGEMA4x. Insertions of the wild-type intron (IS+I) or the ΔORF derivative (IS+ΔI) are indicated. D indicates hybridization signals from pKG2.5 and pKG2.5x DNA and the asterisk (*) a DNA fragment partially digested presumably containing ISb1. (B) Distribution of intron RmInt1 and the newly acquired intron copies (wt or ΔORF) within the 1021 genome. Schematic shows chromosome, pSymA and pSymB with indication of their replication origin, localization of RmInt1 targets, resident intron copies and new insertions upon acquisition of plasmids pKG2.5 or pKGEMA4 as deduced from the Southern analyses of all the transconjugants according to the S.meliloti 1021 genome annotation in (http://bioinfo.genopole-toulouse.prd.fr/annotation/iANT/bacteria/rhime/). IS targets correspond to the following annotated 1021 genes: SMc02205 (ISc1), SMc00848 (ISc2), SMc02411 (ISc3), SMc01382 (ISc4), SMc03084 (ISc5), SMc03305 (ISc6), SMa3000 (ISa1), SMa3014 (ISa2), SMa3015 (ISa3), SMa3018 (ISa4), SMb21616 (ISb1), SMb21703-5 (ISb2) and ISRm10-1 (SMb21709-10).