Abstract
Alternative splicing increases transcriptome and proteome diversification. Previous analyses aiming at comparing the rate of alternative splicing between different organisms provided contradicting results. These contradicting results were attributed to the fact that both analyses were dependent on the expressed sequence tag (EST) coverage, which varies greatly between the tested organisms. In this study we compare the level of alternative splicing among eight different organisms. By employing an EST independent approach we reveal that the percentage of genes and exons undergoing alternative splicing is higher in vertebrates compared with invertebrates. We also find that alternative exons of the skipping type are flanked by longer introns compared to constitutive ones, whereas alternative 5′ and 3′ splice sites events are generally not. In addition, although the regulation of alternative splicing and sizes of introns and exons have changed during metazoan evolution, intron retention remained the rarest type of alternative splicing, whereas exon skipping is more prevalent and exhibits a slight increase, from invertebrates to vertebrates. The difference in the level of alternative splicing suggests that alternative splicing may contribute greatly to the mammal higher level of phenotypic complexity, and that accumulation of introns confers an evolutionary advantage as it allows increasing the number of alternative splicing forms.
INTRODUCTION
Sequencing of the human and mouse genomes has revealed an unexpectedly low number of protein coding genes (∼25 000 in both species), not substantially higher than the number of protein coding genes in the genome of the nematode (19 000), and less than in rice (∼38 000–40 000) (1–4). Alternative splicing, a mechanism that increases transcriptome and proteome diversification by generating multiple mRNA products from a single gene, was suggested as a possible solution to this paradoxical miscorrelation between the number of genes in an organism's genome and its level of phenotypic complexity. According to this assumption, we would expect to find a higher rate of alternative splicing in vertebrates, and in particular in mammals, relative to less complex organisms with similar, or even higher, number of genes (5,6).
Several recent studies estimated that >60% of human and mouse genes undergo alternative splicing (2,7,8). However, previous analyses aiming to compare the rate of alternative splicing between different organisms have provided conflicting results. Brett et al. (9) in a large-scale expressed sequence tag (EST) analysis, estimated that, across a variety of distinct metazoan organisms as humans and nematodes, the rate of alternative splicing is similar. Contrary to that, Kim et al. (10) estimated a greater amount of alternative splicing in mammals compared with invertebrates. However, in both studies, the methods used for measuring the amount of alternative splicing were shown to be dependent on the extent of EST coverage of the different organisms (11). Kan et al. (12) have suggested that given a sufficient amount of EST coverage, alternative splice patterns may be observed for all genes that undergo splicing; hence, the higher the EST coverage, the higher the level of alternative splicing we expect. Therefore, performing such analysis that is dependent on the EST coverage will yield inconclusive findings.
In this study, we employed a straight-forward method for calculating the percentage of alternatively spliced genes and exons in different organisms, by detecting alternative splicing events in gene-oriented clusters of mRNAs and ESTs. The clusters we used were derived from the UniGene database (13). Our analysis included eight eukaryotic organisms that represent distinct evolutionary lineages: flowering plants (Arabidopsis thaliana); nematodes (Caenorhabditis elegans); insects (Drosophila melanogaster, fly); primitive chordates (Ciona intestinalis, sea squirt); and two classes of vertebrates: aves (Gallus gallus, chicken) and mammals (Mus musculus, mouse; Rattus norvegicus, rat; Homo sapiens, human). Supplementary Figure S1 shows the relationship and times of divergence of these eight species. These species were chosen for their relative high coverage of ESTs and mRNAs in UniGene and the advanced state of their genome project. We provide evidence that our results are not dependent on the extent of EST coverage of the different species. Our findings suggest that vertebrates have a substantially higher percentage of alternatively spliced genes compared with other species. We show that exon skipping is, in general, the most prevalent form of alternative splicing in metazoans, while in the single plant we analyzed it is the rarest. We also demonstrated that longer introns flank alternatively-skipped exons compared to constitutive exons and exons with alternative 5′ and 3′ splice sites (5′ss and 3′ss).
MATERIALS AND METHODS
Data preparation
UniGene builds for the eight species were downloaded from the UniGene FTP site. Sea squirt (C.intestinalis) genome was downloaded from the DOE Joint Genome Institute, and the genome sequences of the remaining seven species were downloaded from NCBI's genomes FTP site on December 12, 2005.
Description of the algorithm
Each UniGene cluster has a representative sequence (termed ‘unique sequence’, usually an mRNA), which is the sequence in the cluster with the longest region of high-quality data. Each cluster's unique sequence was blasted against the organism's genome, in order to obtain the genomic localization of the gene represented by the cluster. Only clusters whose unique sequence best blast result had an E-value < 10−5 were considered for further analysis. Then, for each cluster, we built a 200 000 bp contig, composed of a concatenation of 100 000 bp from both sides of the start point of the best blast result. All sequences in the cluster were then aligned to the 200 000 bp contig, taking into account consensus splice signals and removing poly(A) sequences, using sim4 (14). This provided us with the exon–exon junctions of the mRNAs and ESTs in each cluster, and the sizes of exons and introns.
We decided to include only internal exons in our analysis because the prediction of terminal exons using EST alignments is problematic due to the low quality of sequencing at the ends of ESTs (15). For the same reason, we removed internal exons whose start point was <50 nt from the 5′ end of the sequence or whose end point was <50 nt from its 3′ end. As a result, all sequences with less than three exons were discarded (these were mainly ESTs). Therefore, our analysis did not include genes containing only two exons, or intronless genes. We further filtered out some more sequences, according to the following criteria:
Sequences with possible intronic contamination. Intronic contamination was assumed in cases where sim4 detected an exon in an mRNA/EST, whose start point, relative to the genomic sequence, was more than 1 nt from the end point of its preceding exon.
Possible chimeras. Sim4 provides the orientation of each exon relative to the genomic sequence. Sequences containing exons with opposite orientations or with undetermined orientations were considered as putative chimeras and were thus discarded.
Poor sequencing quality. This included sequences whose terminal exons (after removing the original terminal exons) had a sim4 alignment score of <96%.
Sequences that are derived from cancer tissues. Sequences that are termed as derived from a ‘neoplasia’ tissue were discarded, as cancer can increase the amount of aberrant splicing (16).
RefSeq sequences. RefSeq sequences were removed since they are either predicted (XPs) and hence unreliable, or derived from an already submitted sequence (NMs) and could therefore bias the dataset.
We then implemented an algorithm whose input is all exon boundaries of all sequences in a given cluster (in other words, sim4 output). The output of this algorithm is the type of each exon (constitutive or alternative of four possible types) and the number of sequences that support each type. See Supplementary Figure S2 for an example of the algorithm's output for a hypothetical cluster.
Extraction of introns flanking constitutive and alternative exons
We have compiled datasets of exon triplets, consisting of three exons, separated by two introns. These triplets were compiled such that the central exon was the subject of the analysis, while the two flanking exons were always constitutive. This way, we gathered four different datasets, in which the center exon is either constitutive, skipped, alternative 3′ss or alternative 5′ss. The human constitutive, skipped, alternative 3′ss and alternative 5′ss datasets consisted of 56 693, 2894, 2002 and 1810 exons, respectively. Similarly, the four mouse datasets consisted of 60 568, 1891, 1922 and 1804 exons. The four rat datasets consisted of 41 074, 540, 531 and 548 exons. The four chicken datasets consisted of 21 212, 403, 458 and 432 exons. The four Ciona datasets consisted of 22 591, 169, 239 and 211 exons. The four Drosophila datasets consisted of 6215, 124, 82 and 71 exons. The four C.elegans datasets consisted of 8247, 73, 71 and 75 exons. Finally, the four Arabidopsis datasets consisted of 24 721, 39, 214 and 231 exons.
RESULTS
Detection of alternative splicing events in UniGene clusters
UniGene is a system for automatically partitioning GenBank sequences (ESTs and mRNAs) into a non-redundant set of gene-oriented clusters. In order to detect evidence for alternative splicing events in genes that are represented by UniGene clusters, we downloaded UniGene builds of eight eukaryotic species, and their genome sequences.
Detailed description of the algorithm that detects alternative splicing events in UniGene clusters is presented in Materials and Methods. In brief, we aligned the sequences in the UniGene clusters to their corresponding genomic loci, and identified exons according to the consensus splice signals that flanked them. We then discarded sequences from cancer tissues, as these can increase the amount of aberrant splicing (16), and RefSeq sequences (17), as they are either predicted (XPs), or derived from already submitted sequences (NMs) and hence bias the dataset. The algorithm provides the type of alternative splicing that each alternatively spliced exon undergoes (Supplementary Figure S2 illustrates an output of the algorithm for a hypothetical cluster).
Detection of alternative splicing depends on EST coverage
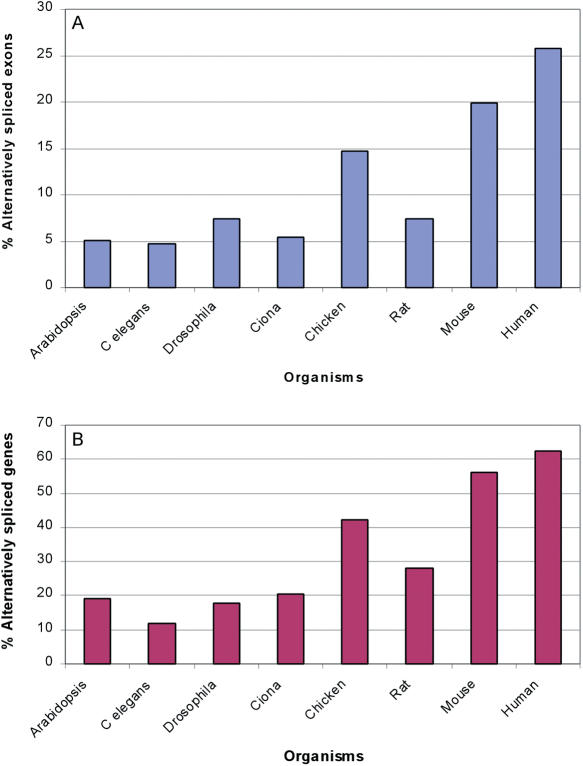
We have implemented the algorithm on UniGene clusters of eight different organisms (Human, Mouse, Rat, Chicken, Ciona, Drosophila, C.elegans and Arabidopsis) and calculated the percentage of exons that undergo alternative splicing, as well as the percentage of clusters, in which one or more exons are alternatively spliced. In this analysis, we have included all mRNA and EST sequences available (except from cancer and RefSeq). Thus, the results are dependent on the EST coverage. Indeed, we find that human and mouse, which exhibit a relatively high-EST coverage, demonstrate a significantly higher amount of detectable alternative exons, as well as alternative clusters, compared with the other organisms (Figure 1). The effect of the EST coverage is clearly demonstrated by the results of mouse and rat. Although these two organisms are closely related [23 million years of divergence (18)], and hence we could expect similar levels of alternative splicing, we detect a significantly higher level of alternative splicing events in mouse (P ≈ 0, χ2 = 7016.96, df = 1), which is compatible with the huge difference in the EST coverage. Thus, as expected, the amount of detectable alternative splicing depends on the EST coverage, and hence comparing the amount of alternative splicing between organisms without regarding the different EST coverage might lead to erroneous conclusions.
Figure 1.
EST-dependent analysis of the levels of alternatively spliced genes and exons among eukaryotes. Percentage of alternatively spliced exons (A) and genes (B) among eight different organisms. The percentages were calculated based on the analysis of UniGene clusters using all reliable data available.
Normalization of the EST coverage
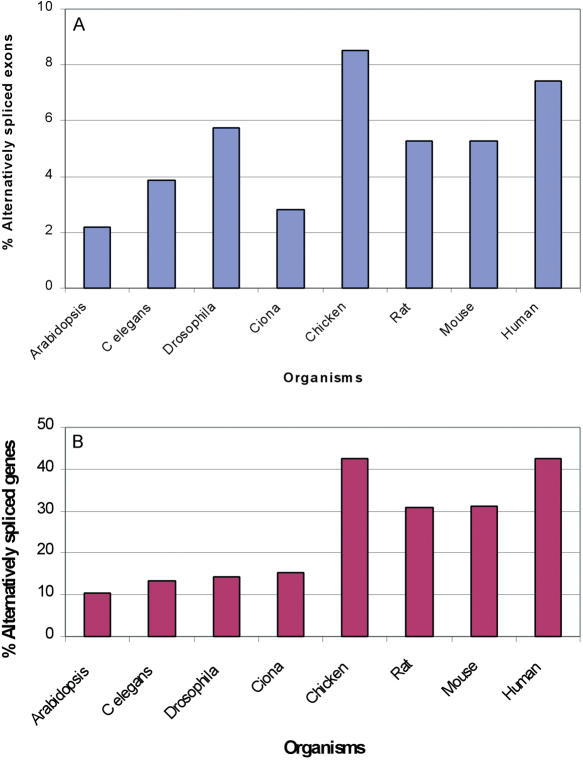
In order to compare the extent of alternative splicing between different organisms, one must try to overcome the bias introduced by the different EST coverage of each of the organisms. To create comparable datasets for each organism, we decided to employ a similar approach to the one utilized by Brett et al. (9), in which we randomly selected ESTs for each organism. Here, we required the final number of ESTs in the compared organisms' genes to be the same. To this end, we extracted UniGene clusters consisting of at least 10 sequences. For each of the clusters, we then randomly selected 10 ESTs, and searched for evidence of alternative splicing. We repeated this randomization process for 100 times, so the final number of alternative splicing events is the average of the 100 repeated calculations. In this analysis we discarded mRNA sequences, and used only non-cancerous EST data, since mRNAs are usually longer and of higher quality, and therefore different fractions of mRNAs versus ESTs in the different organisms could introduce a bias to the results. The results clearly show that different organisms have different levels of alternative exons, as well as alternatively spliced genes, and that mouse and rat exhibit comparable levels of alternatively spliced genes and exons despite the differences in the EST coverage (Figure 2).
Figure 2.
Minimization of the dependence on EST coverage by randomizations. Percentage of alternatively spliced exons (A) and genes (B) among eight different organisms. Percentages were calculated based on the analysis of UniGene clusters, after removing mRNA data. Ten ESTs were randomly selected and this analysis repeated 100 times for each cluster.
Additional filtration of ESTs
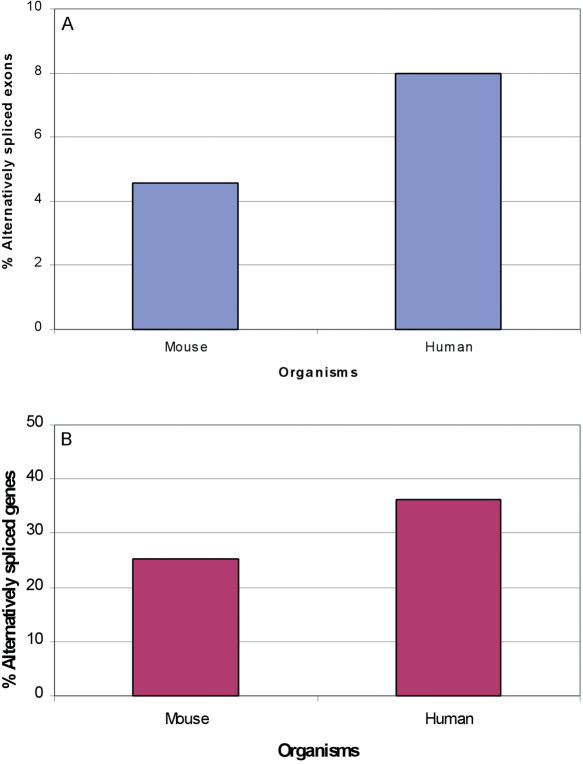
The above randomization process ensures that each cluster is supported by the same number of ESTs. However, we were concerned that the varying quality of ESTs and different methods of construction could introduce another bias to the results. In order to further validate that different organisms indeed exhibit different levels of alternative exons and genes, we first decided to compare similar sets of genes. To this end, we used the HomoloGene database (13) to extract sets of human–mouse homologs, and extracted the corresponding UniGene clusters. To make sure that EST sequence errors or different sequencing methods are not the source for the differences in the alternative splicing patterns, we decided to also compare similar sets of ESTs. We therefore applied an additional filter on the EST sequences, remaining only with ESTs derived from the Mammalian Gene Collection (MGC) project, which are considered to be of high quality and were produced by the same technique (19). The human and mouse organisms were selected since they are the only ones having sufficient MGC EST data. We analyzed clusters where both human and mouse had at least 10 MGC ESTs, and repeated the above randomization process on this subset of clusters. The results reveal that human has a significantly higher amount of alternatively spliced genes (P < 1E − 5, χ2 = 28.82, df = 1) as well as alternative exons (p < 1E − 5, χ2 = 73.2, df = 1), compared with mouse (Figure 3).
Figure 3.
Use of homologous clusters and MGC ESTs. Percentage of alternatively spliced exons (A) and genes (B) among eight different organisms. Percentages were calculated based on the analysis of UniGene clusters, using only ESTs derived from the MGC project. The analysis was performed on homologous human/mouse clusters, where both contained at least 10 ESTs, and 10 ESTs were repeatedly randomly selected.
Alternative splicing in chicken
Our results indicate that chicken has a high rate of alternative splicing, compared with the other tested organisms. We found no support for this finding in the literature. There is a possibility, therefore, that these results are artifacts as a result of low-quality ESTs or genome sequencing errors. Nevertheless, the distribution of the alternative exons in the four types of alternative splicing pattern is similar to the one found in other vertebrates (Figure 4). Hence, there is probably no single type of alternative splicing that contaminates the dataset. Low-quality ESTs might also introduce spurious alternative exon events, as a result of insertion of few nucleotides. Examination of the exons that undergo exon skipping revealed that all are longer than 10 nt and almost 90% are longer than 50 nt, reducing the likelihood of such possibility. We next looked for EST libraries, which support an unusual number of alternative splicing events relative to their size, and hence are potentially low-quality libraries that might cause such an artifact. From the 140 different chicken libraries we found one such library, with a somewhat unusual number of ESTs supporting alternative splicing. However, discarding it from the analysis did not alter the results significantly (data not shown). We also confirmed that the high level of alternative splicing is not a result of many ESTs that are derived from embryo EST libraries, or high level of ESTs generated from specific tissues/cells, which might alter the results (data not shown). Overall, we found no evidence that the elevated rates of alternative splicing are artifacts, although further examination is required. This suggests that vertebrates have higher levels of alternative splicing compared with invertebrates.
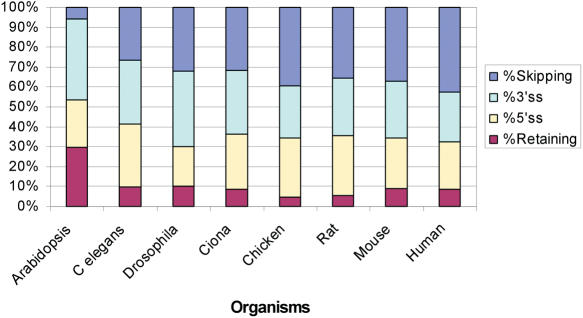
Figure 4.
Relative prevalence of the different types of alternatively spliced exons in different species. The relative prevalence of the four types of alternatively spliced exons (exon skipping, alternative 3′ss, alternative 5′ss and intron retention) are shown for the different organisms.
Prevalence of the four types of alternative splicing in different species
The average size of introns is known to be highly variable among eukaryotes, with mammalian introns having the largest average size (2). Exons are much more uniform in size, however exons in fly and nematode are known to be longer, compared with mammalian exons (2, 20). The results of our analysis are consistent with these observations (data not shown). We therefore wanted to examine whether the exon/intron changes during evolution, which were accompanied by changes in the regulation of alternative splicing (21), have led to differences in the relative prevalence of the four main types of alternative splicing that exons can undergo: exon skipping, alternative 5′ss and 3′ss, and intron retention [reviewed in (22)]. It was already reported that, among conserved alternative splicing events between human and mouse, exon skipping is the most prevalent type of alternative splicing and intron retention is the rarest (23). Our results reveal a similar trend, in which intron retention is the rarest form of alternative splicing in all seven metazoan species examined in our analysis, while exon skipping is the most prevalent form in vertebrates (but not in invertebrates) (Figure 4). The plant differs from metazoans in this respect, with a high level of intron retention events, and with exon skipping being the rarest form of alternative splicing. High levels of intron retention in A.thaliana were already reported (24,25). Overall, our results indicate that over the course of metazoan evolution, the prevalence of the four types of alternative splicing remained similar, with exon skipping exhibiting a slight increase, from invertebrates to vertebrates.
Lengths of introns flanking alternative and constitutive exons
Hertel and co-workers (26) suggested that since splice-site recognition is less efficient across the exon (exon definition) alternative splicing is more likely to occur in exons flanked by long introns. Supporting this assumption, they found that alternatively skipped exons in Drosophila are flanked by significantly longer introns, a phenomenon that was observed in human as well, but with less significant results. We have compiled datasets of exon triplets, namely constitutive and alternatively skipped exons flanked by two constitutive exons, for each of the eight organisms (see Materials and Methods). We then extracted the flanking introns of each of the constitutive and alternative exons and calculated their lengths (Table 1). The results reveal that, in general, alternatively skipped exons are indeed flanked by longer introns than constitutive ones. We repeated this analysis with alternative 5′ss and 3′ss exons. Interestingly, in almost all cases, lengths of introns flanking these alternative exons were not longer than introns flanking constitutive exons. These results suggest that intron length plays a key role in the different types of alternative splicing.
Table 1.
Lengths of introns flanking constitutively and alternatively spliced exons
| Organism | Constitutive | Alternative 3′ss | Alternative 5′ss | Skipping |
|---|---|---|---|---|
| Upstream | ||||
| Human | 2857.9 | 2907.9* | 3079.7a | 3664.7*** |
| Mouse | 2446.9 | 2532.5* | 2853.0a | 3461.4*** |
| Rat | 2562.1 | 2225.8a | 2271.5a | 2926.2a |
| Chicken | 1866.1 | 1960.7a | 1681.7a | 2925.6** |
| Ciona | 619.0 | 528.5a | 1318.4a | 800.2a |
| Drosophila | 1024.1 | 1541.0a | 1442.0a | 2816.6** |
| C.elegans | 538.8 | 680.2a | 313.6a | 866.3a |
| Arabidopsis | 148.7 | 144.2a | 142.0a | 224.7* |
| Downstream | ||||
| Human | 2725.4 | 2600.0a | 2782.7a | 3417.7*** |
| Mouse | 2349.6 | 2320.9a | 2453.4* | 3117.8*** |
| Rat | 2436.6 | 1913.3a | 2157.1a | 2701.3a |
| Chicken | 1759.0 | 1855.2a | 1693.7a | 2227.6* |
| Ciona | 641.2 | 521.8a | 665.8a | 950.9a |
| Drosophila | 799.3 | 1202.8a | 2135.3* | 2514.1** |
| C.elegans | 557.2 | 426.0a | 822.4a | 1359.2* |
| Arabidopsis | 147.7 | 141.4a | 129.0a | 155.9a |
Average length of upstream and downstream introns flanking constitutive, alternative 3′ss, alternative 5′ss and skipped exons, are shown. In order to determine whether introns flanking alternative exons are longer than introns flanking constitutive exons, the averages of introns flanking alternative 3′ss, alternative 5′ss and skipped exons were compared to introns flanking constitutive exons. *P < 0.05; **P < 0.01; ***P< 1E−5.
aInsignificant.
DISCUSSION
In this article we address the issue of whether different eukaryotes have different levels of alternatively spliced exons and genes. The results we present emphasize that the differences in the EST coverage between different organisms lead to an unreliable comparison of the level of alternative splicing. Moreover, we argue that not only the extent of the EST coverage, but also the quality of ESTs and the different methods, by which they were constructed, may introduce a bias to the results. Hence, it is impractical to determine the exact level of alternative exons and genes in the tested organisms. However, by using normalized reliable subsets of ESTs that support homologous genes, we confirmed that the relative level of alternative splicing is not constant among eukaryotes. Namely, humans have higher rates of alternative exons and genes than mice. Moreover, we found that the results for human and mouse using the MGC reliable subset of ESTs were, in general, similar to the results we revealed using the random samples of the entire EST dataset. We also found that although rat has a much lower coverage of ESTs than mouse, it was found to have similar rates of alternatively spliced genes and exons, as expected for such closely related organisms. Hence, these results suggest that the analysis that was conducted using random samples of the entire EST dataset is reliable, and that the possible effect of different EST quality and methods of construction are probably negligible.
Two recent analyses that compared the level of alternative splicing among different organisms had contradicting results. Kim et al. (10) in a somewhat indirect method to calculate the level of alternative splicing, revealed that the level of alternative splicing varies between different organisms. However, in reply, Harrington et al. (11) found that this method was dependent on the EST coverage of the organisms. Brett et al. (9) in an analysis conducted on a random set of 650 mRNAs, which were aligned to 100 000 ESTs, found that the level of alternative splicing events remained constant across a variety of distinct metazoan organisms. Although this method is similar to the one presented here, we aligned the entire EST dataset to the genome, and examined only bona fide alternative splicing events exhibiting canonical splice sites. We further required at least 10 ESTs to support a gene. We also discarded terminal exons from the analysis, because of the low quality of sequencing at the ends of ESTs, and also ESTs from cancer tissues, since cancer can increase the amount of aberrant splicing. We then further validated the applicability of this approach using a comparable dataset of orthologous genes for human and mouse, and identical numbers of high-quality ESTs, which were generated by the same methods that eliminate potential data biases.
The different levels of alternative splicing between vertebrates and invertebrates suggest that the appearance of vertebrates, some 300 million years ago, was accompanied by an increase in the number of alternatively spliced genes. However, the sea squirt shows an exception from the correlation between the gradual rise in the percentage of alternatively spliced genes during evolution and the order of appearance of the different species in evolution. This organism exhibits a percentage of alternatively spliced genes lower than that of invertebrates (Figure 2), though it is located closer to vertebrates on the evolutionary tree (Supplementary Figure S1). Nevertheless, recent studies support that the Ciona genome may not be a good representative of the ancestral Chordate genome, as it suffered from major loss of genes and introns (27,28). Sea squirt free-swimming larvae metamorphose into sessile adults in a very early stage of its life cycle (29). As its percentage of alternatively spliced genes and exons is closer to that of A.thaliana than to that of the other metazoans, it may be that evolution dictates a lower rate of alternative splicing in sessile organisms. In the same manner, chicken exhibits higher levels of alternative exons and genes, and Drosophila and C.elegans exhibits higher levels of alternative exons, but not alternative genes, which is somewhat unexpected (Figure 2). In Drosophila and C.elegans, this high level of alternative exons was found to be attributed to a relatively low number of exons per gene. In chicken, however, the reason is unclear. There is no evidence in the literature for such an elevated level of alternative splicing in chicken, still we could not find evidence that these results might be erroneous. Although further examination is required to determine if avians contain high levels of alternative splicing, these results suggest that a major increase in the level of alternative splicing occurred in the transition from invertebrates to vertebrates.
Our findings also indicate that over the course of metazoan evolution, intron retention has remained the rarest type of alternative splicing, and exon skipping is usually the more prevalent one (Figure 4). The long introns in mammals and especially in humans, relative to other species, could lead to suboptimal recognition of exons by the basal splicing machinery and result in higher levels of alternative splicing. This is supported by the finding that skipped exons are flanked by longer introns, and also by the finding that mammals (and especially humans) have higher fractions of exons that undergo skipping. However, since exon skipping is prevalent in all metazoans, intron size may not be the single factor that governs this mechanism.
Compared with metazoans, the plant, A.thaliana, has a very low percentage of exon skipping events and a high level of intron retention (Figure 4), and also exhibits unique introns which are usually short in length and U-rich (30). Interestingly, among unicellular eukaryotes, which generally exhibit very low rates of alternative splicing, intron retention is the most frequent alternative splicing event (22). These findings may suggest that intron retention is the most ancient form of alternative splicing that appeared in evolution before the emergence of the exon definition mechanism in higher eukaryotes, which enabled exon skipping events [see also (22)]. Therefore, the high level of intron retention in A.thaliana may reflect a lower rate of evolution of the alternative splicing mechanism in plants after their divergence from animals (∼1.5 billion years ago). As plants exhibit a very high level of polyploidy (31), it may be that gene duplication, another mechanism for generating genomic diversity, plays a greater role in increasing transcriptome and proteome complexity in plants compared to metazoans, and compensates for the need of alternative splicing (32).
Finally, in order for the analysis to be reliable and independent of the EST coverage, quality and method of construction, we concentrated on highly reliable homogenous subsets of ESTs and on homologous genes. The intensive filtration process we utilized most probably results in an underestimation of the real rate of alternative splicing, still we provide reliable evidence that the rate of alternative splicing is not constant among eukaryotes.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
We would like to thank M. Amit, A. Goren and E. Koren for reading the manuscript. This work was supported by a grant from the Israel Science Foundation (1449/04 and 40/05), MOP Germany–Israel, GIF, ICA through the Ber–Lehmsdorf Memorial Fund, and DIP. E.K. is a fellow of the Edmond J. Safra Bioinformatics Program at Tel-Aviv University. E.K. is funded in part by EURASNET. Funding to pay the Open Access publication charges for this article was provided by EURASNET.
Conflict of interest statement. None declared.
REFERENCES
- 1.Consortium C.e.S. Genome sequence of the nematode C.elegans: a platform for investigating biology. Science. 1998;282:2012–2018. doi: 10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
- 2.Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 3.Yu J., Wang J., Lin W., Li S., Li H., Zhou J., Ni P., Dong W., Hu S., Zeng C., et al. The Genomes of Oryza sativa: a history of duplications. PLoS Biol. 2005;3:e38. doi: 10.1371/journal.pbio.0030038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Waterston R.H., Lindblad-Toh K., Birney E., Rogers J., Abril J.F., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 5.Modrek B., Lee C. A genomic view of alternative splicing. Nature Genet. 2002;30:13–19. doi: 10.1038/ng0102-13. [DOI] [PubMed] [Google Scholar]
- 6.Graveley B.R. Alternative splicing: increasing diversity in the proteomic world. Trends Genet. 2001;17:100–107. doi: 10.1016/s0168-9525(00)02176-4. [DOI] [PubMed] [Google Scholar]
- 7.Sharov A.A., Dudekula D.B., Ko M.S. Genome-wide assembly and analysis of alternative transcripts in mouse. Genome Res. 2005;15:748–754. doi: 10.1101/gr.3269805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johnson J.M., Castle J., Garrett-Engele P., Kan Z., Loerch P.M., Armour C.D., Santos R., Schadt E.E., Stoughton R., Shoemaker D.D. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science. 2003;302:2141–2144. doi: 10.1126/science.1090100. [DOI] [PubMed] [Google Scholar]
- 9.Brett D., Pospisil H., Valcarcel J., Reich J., Bork P. Alternative splicing and genome complexity. Nature Genet. 2002;30:29–30. doi: 10.1038/ng803. [DOI] [PubMed] [Google Scholar]
- 10.Kim H., Klein R., Majewski J., Ott J. Estimating rates of alternative splicing in mammals and invertebrates. Nature Genet. 2004;36:915. doi: 10.1038/ng0904-915. 916; author reply 916–917. [DOI] [PubMed] [Google Scholar]
- 11.Harrington E.D., Boue S., Valcarcel J., Reich J.G., Bork P. Estimating rates of alternative splicing in mammals and invertebrates. Nature Genet. 2004;36:915–916. doi: 10.1038/ng0904-915. author reply 916–917. [DOI] [PubMed] [Google Scholar]
- 12.Kan Z., States D., Gish W. Selecting for functional alternative splices in ESTs. Genome Res. 2002;12:1837–1845. doi: 10.1101/gr.764102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wheeler D.L., Barrett T., Benson D.A., Bryant S.H., Canese K., Church D.M., DiCuccio M., Edgar R., Federhen S., Helmberg W., et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2005;33:D39–D45. doi: 10.1093/nar/gki062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Florea L., Hartzell G., Zhang Z., Rubin G.M., Miller W. A computer program for aligning a cDNA sequence with a genomic DNA sequence. Genome Res. 1998;8:967–974. doi: 10.1101/gr.8.9.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sorek R., Ast G., Graur D. Alu-containing exons are alternatively spliced. Genome Res. 2002;12:1060–1067. doi: 10.1101/gr.229302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Z., Lo H.S., Yang H., Gere S., Hu Y., Buetow K.H., Lee M.P. Computational analysis and experimental validation of tumor-associated alternative RNA splicing in human cancer. Cancer Res. 2003;63:655–657. [PubMed] [Google Scholar]
- 17.Pruitt K.D., Tatusova T., Maglott D.R. NCBI Reference Sequence (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2005;33:D501–D504. doi: 10.1093/nar/gki025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Adkins R.M., Gelke E.L., Rowe D., Honeycutt R.L. Molecular phylogeny and divergence time estimates for major rodent groups: evidence from multiple genes. Mol. Biol. Evol. 2001;18:777–791. doi: 10.1093/oxfordjournals.molbev.a003860. [DOI] [PubMed] [Google Scholar]
- 19.Gerhard D.S., Wagner L., Feingold E.A., Shenmen C.M., Grouse L.H., Schuler G., Klein S.L., Old S., Rasooly R., Good P., et al. The status, quality, and expansion of the NIH full-length cDNA project: the Mammalian Gene Collection (MGC) Genome Res. 2004;14:2121–2127. doi: 10.1101/gr.2596504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lynch M., Kewalramani A. Messenger RNA surveillance and the evolutionary proliferation of introns. Mol. Biol. Evol. 2003;20:563–571. doi: 10.1093/molbev/msg068. [DOI] [PubMed] [Google Scholar]
- 21.Yeo G., Hoon S., Venkatesh B., Burge C.B. Variation in sequence and organization of splicing regulatory elements in vertebrate genes. Proc. Natl Acad. Sci. USA. 2004;101:15700–15705. doi: 10.1073/pnas.0404901101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ast G. How did alternative splicing evolve? Nature Rev. Genet. 2004;5:773–782. doi: 10.1038/nrg1451. [DOI] [PubMed] [Google Scholar]
- 23.Sugnet C.W., Kent W.J., Ares M., Jr, Haussler D. Transcriptome and genome conservation of alternative splicing events in humans and mice. Pac. Symp. Biocomput. 2004;2004:66–77. doi: 10.1142/9789812704856_0007. [DOI] [PubMed] [Google Scholar]
- 24.Ner-Gaon H., Halachmi R., Savaldi-Goldstein S., Rubin E., Ophir R., Fluhr R. Intron retention is a major phenomenon in alternative splicing in Arabidopsis. Plant J. 2004;39:877–885. doi: 10.1111/j.1365-313X.2004.02172.x. [DOI] [PubMed] [Google Scholar]
- 25.Iida K., Seki M., Sakurai T., Satou M., Akiyama K., Toyoda T., Konagaya A., Shinozaki K. Genome-wide analysis of alternative pre-mRNA splicing in Arabidopsis thaliana based on full-length cDNA sequences. Nucleic Acids Res. 2004;32:5096–5103. doi: 10.1093/nar/gkh845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fox-Walsh K.L., Dou Y., Lam B.J., Hung S.P., Baldi P.F., Hertel K.J. The architecture of pre-mRNAs affects mechanisms of splice-site pairing. Proc. Natl Acad. Sci. USA. 2005;102:16176–16181. doi: 10.1073/pnas.0508489102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hughes A.L., Friedman R. Loss of ancestral genes in the genomic evolution of Ciona intestinalis. Evol. Dev. 2005;7:196–200. doi: 10.1111/j.1525-142X.2005.05022.x. [DOI] [PubMed] [Google Scholar]
- 28.Raible F., Tessmar-Raible K., Osoegawa K., Wincker P., Jubin C., Balavoine G., Ferrier D., Benes V., de Jong P., Weissenbach J., et al. Vertebrate-type intron-rich genes in the marine annelid Platynereis dumerilii. Science. 2005;310:1325–1326. doi: 10.1126/science.1119089. [DOI] [PubMed] [Google Scholar]
- 29.Satou Y., Takatori N., Fujiwara S., Nishikata T., Saiga H., Kusakabe T., Shin-i T., Kohara Y., Satoh N. Ciona intestinalis cDNA projects: expressed sequence tag analyses and gene expression profiles during embryogenesis. Gene. 2002;287:83–96. doi: 10.1016/s0378-1119(01)00826-5. [DOI] [PubMed] [Google Scholar]
- 30.Wang B.B., Brendel V. Genome-wide comparative analysis of alternative splicing in plants. Proc. Natl Acad. Sci. USA. 2006;103:7175–7180. doi: 10.1073/pnas.0602039103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Adams K.L., Wendel J.F. Polyploidy and genome evolution in plants. Curr. Opin. Plant Biol. 2005;8:135–141. doi: 10.1016/j.pbi.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 32.Kopelman N.M., Lancet D., Yanai I. Alternative splicing and gene duplication are inversely correlated evolutionary mechanisms. Nature Genet. 2005;37:588–589. doi: 10.1038/ng1575. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.