Abstract
RNA interference (RNAi) can be induced in vitro either by application of synthetic short interfering RNAs (siRNAs), or by intracellular expression of siRNAs or short hairpin RNAs (shRNAs) from transfected vectors. The most widely used promoters for siRNA/shRNA expression are based on polymerase III (Pol III)-dependent transcription. We developed an alternative vector for siRNA/shRNA expression, using a mouse RNA polymerase I (Pol I) promoter. Pol I-dependent transcription serves in cells for production of ribosomal RNA (rRNA), and as such, is ubiquitously and stably active in different cell types. As Pol I-dependent transcription is highly species-specific, Pol I-based system provides an important biosafety advantage with respect to silencing of genes with unknown functions.
INTRODUCTION
RNA interference (RNAi) is a post-transcriptional gene-silencing mechanism, by which a specific degradation of mRNA is induced by homologous double-stranded RNAs (dsRNAs). Since its discovery, RNAi has rapidly evolved into a powerful technique to knock-down gene expression and gained ground as an indispensable tool to study gene function both in vitro and in vivo [reviewed in (1–3)]. In plants and animals, such as Caenorhabditis elegans and Drosophila, RNAi can be activated by long dsRNAs, which are processed by an intracellular machinery to short interfering RNAs (siRNAs) [(4–7); reviewed in (8)].
However, in mammalian cells long dsRNAs cause general translational inhibition and unspecific RNA degradation (9,10). These effects can be avoided by introducing in vitro-synthesized siRNAs into the cells (9,10). The resulting silencing is very pronounced, in particular due to the high siRNAs transfection efficiency, greatly exceeding the one for DNA vectors. Therefore, this technique has been widely adopted in experimental paradigms, such as pathway profiling and drug screening [reviewed in (11,12)]. Its major drawback, however, resides in the transient nature of silencing. Studies of gene function often require the establishment of cell lines or transgenic organisms, in which siRNAs should be expressed from stably integrated cassettes. In expression vectors, siRNAs are normally processed from a short hairpin RNA (shRNA) ∼70 nt in length, structurally similar to naturally occurring microRNA (miRNA) precursor. This sequence should fulfil strict length requirements to result in an effective siRNA. Transcripts generated from conventional RNA polymerase II (Pol II) promoter-based vectors are therefore not suitable for such a purpose, as they undergo extensive post-transcriptional modifications, such as capping and poly(A) tailing (10). To circumvent this problem, RNA polymerase III (Pol III) promoter-driven cassettes were developed (13–16). Recently, the use of a cytomegalovirus (CMV) promoter to generate siRNAs was made possible by insertion of a human miRNA miR-30 precursor and neighboring genomic sequences into the expression vector (17). Here, we present an alternative method for expression of siRNAs, using ribosomal transcription units driven by RNA polymerase I (Pol I) promoters. Similarly to Pol III-driven transcription, Pol I-mediated transcription is very robust, ubiquitous, devoid of extensive RNA modifications and is characterized by clearly defined transcription start and termination sites (18). In addition, it has no limits for transcript size, and even more importantly, possesses a high species specificity (19). This latter feature, which is unique among all RNA polymerases, provides an interesting potential regarding biosafety of experiments.
MATERIALS AND METHODS
Cloning
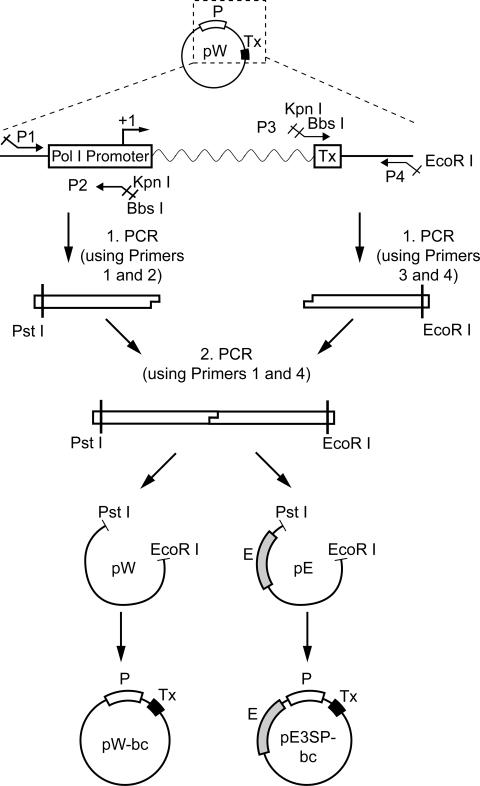
All Pol I-containing plasmids described here were derived from the ribosomal minigenes pW (20), kindly provided by Dr J. Cavaille, Université Paul Sabatier, Toulouse, France, and pE3SP (21). Part of the pW vector was amplified in two steps using primers P1–P4 (Figure 1). All primers were synthesized by MWG Biotech. Primer sequences were as follows: P1 (M13r-based): 5′-CAATTTCACACAGGAAACAGCTATGACC-3′; P2: 5′-ATGTCTTCGAAGTCCATGGTACCTATCTCCAGGTC-3′; P3: 5′-ACCATGGACTTCGAAGACTATCCCCCCCAACTTCG-3′; P4 (M13f-based): 5′-GTCACGACGTTGTAAAACGACGGCCAGT-3′. As a first step, the distal arms of the construct were PCR amplified using P1 and P2, or P3 and P4, respectively. PCR of a total volume of 50 μl were carried out, with each PCR containing 200 μM each dNTP (Roche), 1 μM of each primer, 100 ng of plasmid pW and 2 U of Taq polymerase (Roche). Thermocycling began with 5 min at 95°C followed by 2 cycles of 45 s at 95°C, 60 s at 50°C and 45 s at 72°C, followed by 18 cycles of 45 s at 95°C, 60 s at 60°C and 45 s at 72°C. PCR products were purified with the QiaQuick Gel Extraction Kit (Qiagen) according to the manufacturer's protocol and dissolved in 40 μl of elution buffer. Both arms of the construct were joined in a PCR using primers P1 and P4. First, the annealing reaction of a total volume of 100 μl was carried out, using 200 μM of each dNTP, 1 μl of each product from the first step PCR (P1–P2 and P3–P4, respectively) and 3 U of Taq polymerase. The annealing reaction required five cycles of thermocycling, each of 60 s at 95°C, 2 min at 50°C and 60 s at 72°C. Subsequently, primers (P1 and P4, 1 μM each) were added, and the PCR was carried out for 20 cycles of 45 s at 95°C, 60 s at 60°C and 45 s at 72°C. PCR products were digested with PstI and EcoRI (restriction sites were incorporated into the P1 and P4 primers, respectively), and subcloned either into the vector pW or pE3SP, also digested with PstI and EcoRI, resulting in pW-bc and pE3SP-bc (‘bc’ standing for ‘basic construct’). To generate the pW-MCS (multiple cloning site) and pE3SP-MCS plasmids, oligonucleotides MCS.s and MCS.as were annealed and subcloned into BbsI/KpnI digested pW-bc and pE3SP-bc, respectively. Oligonucleotide sequences were as follows: MCS.s: 5′-GTCTTCGAAGATGTCGAGCTCTAGATATCAATTGGTACCATGGGAAGACTA-3′; MCS.as: 5′-GGGATAGTCTTCCCATGGTACCAATTGATATCTAGAGCTCGAGATCTTCGAAGACGTAC-3′.
Figure 1.
Construction of the vectors pW-bc and pE3SP-bc. Part of the pW ribosomal minigene was PCR amplified in two steps. First, two fragments were generated using primers P1 and P2, or P3 and P4, respectively. Second, both fragments were joined by PCR with primers P1 and P4. The restriction sites PstI and EcoRI, introduced through PCR primers P1 and P4, were used to insert the resulting fragment into the pW and pE3SP (pE) vector backbones. P, mouse Pol I promoter; Tx, transcription termination signal; bc, basic construct; E, enhancer.
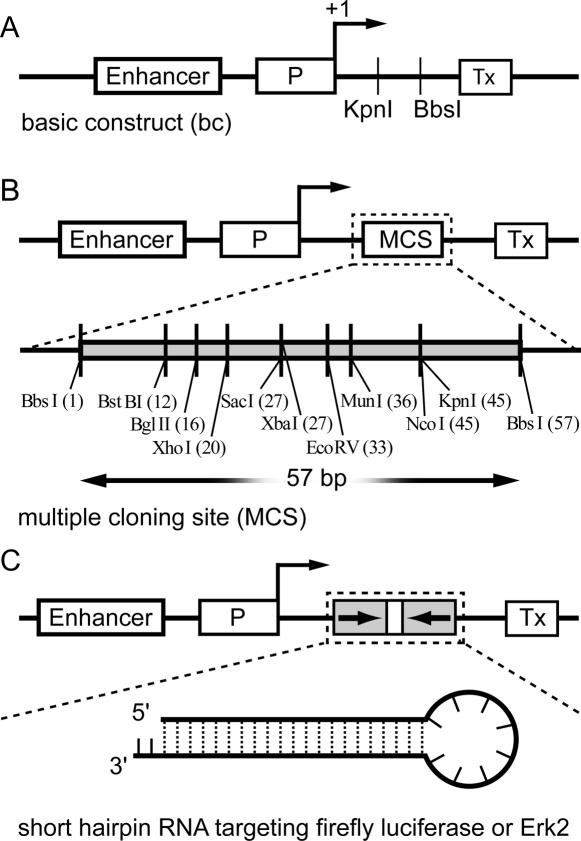
To generate the new RNAi mediating plasmids pW-shLuc and pE3SP-shLuc for knock-down of the firefly luciferase reporter, shLuc-s and shLuc-as oligonucleotides were annealed and subcloned into the BbsI-digested pW-MCS and pE3SP-MCS, respectively (Figure 2). This restriction site lies within the formerly introduced MCS of pW-MCS. Oligonucleotide sequences were as follows: shLuc.s: 5′-AGGTGGATTCCAATTCAGCGGGAGCCACCTGATGAAGCTTGATCGGGTGGCTCTCGCTGAGTTGGAATCCAT-3′; shLuc.as: 5′-GGGAATGGATTCCAACTCAGCGAGAGCCACCCGATCAAGCTTCATCAGGTGGCTCCCGCTGAATTGGAATCC-3′.
Figure 2.
Schematic representation of vectors used in the present study. (A) pE3SP-bc, basic construct vector, containing the enhancer for Pol I-dependent transcription, the sequence of mouse Pol I promoter (P) upstream of transcription start, a transcription termination signal (Tx) and the KpnI and BbsI sites for cloning purposes. (B) pE3SP-MCS vector contains a multiple cloning site for additional cloning versatility. (C) pE3SP-shLuc and pE3SP-Erk2 express short hairpin RNAs targeting firefly luciferase and Erk2, respectively.
Generation of pE3SP-shErk2, which was aimed at silencing Erk2, the endogenous MAP kinase p42, followed the same strategy as for cloning of pE3SP-shLuc. The oligonucleotides shErk2-s and shErk2-as were annealed and also subcloned into the BbsI-digested pE3SP-MCS. Oligonucleotide sequences were as follows: shErk2-s: 5′-AGGTGGAAGATCTGAATTGTATAATAAGAAGCTTGTTATTATACAATTCAGATCTTCCAT-3′; shErk2-as: 5′-GGGAATGGAAGATCTGAATTGTATAATAACAAGCTTCTTATTATACAATTCAGATCTTCC-3′. To have a proper control for the specificity of our RNAi effect on Erk2, we produced a control vector expressing a shRNA that targets an Erk2-unrelated sequence. Thus, pE3SP-shCon was generated following the same cloning strategy as before. Oligonucleotides shCon-s and shCon-as were annealed and subcloned into BbsI-digested pE3SP-MCS. Oligonucleotides were as follows: con-s: 5′-AGGTGTACATGTGTAATAGCTCCTTCAAGAGAGGAGCTATTACACATGTACAT-3′; con-as: 5′-GGGAATGTACATGTGTAATAGCTCCTCTCTTGAAGGAGCTATTACACATGTAC-3′.
Cell culture
FM3A cells were cultured in RPMI 1640 medium with 10% Newborn Serum. HEK293, HT22, HN9, Neuro-2a and HeLa cells were cultured in DMEM (Invitrogen) supplemented with 10% heat-inactivated fetal calf serum (FCS) (Invitrogen), 2 mM glutamine (Invitrogen) and 1% antibiotic-antimycotic mixture (penicillin–streptomycin–amphotericin) (Invitrogen) at 37°C in a humidified 5% CO2 incubator.
IDG3.2 embryonic stem (ES) cells were grown on gelatine-coated tissue culture plates in ES cell medium (DMEM, 15% FCS, 2 mM l-glutamine, 20 mM HEPES, 1 mM sodium pyruvate, non-essential amino acids, 0.1 mM β-mercaptoethanol and 1500 U/ml leukaemia inhibiting factor (Chemicon).
In vitro transcription assays
Nuclear extracts were prepared from exponentially growing FM3A cells. To assay Pol I-specific transcription, 25 μl assays contained 50 ng of template DNA (pE3SP-shLuc), 30 μg of nuclear extract proteins, 12 mM Tris–HCl (pH 7.9), 0.1 mM EDTA, 5 mM MgCl2, 80 mM KCl, 10 mM creatine phosphate, 12% (v/v) glycerol, 0.66 mM each of ATP, GTP and CTP, 0.012 mM UTP and 0.1 μCi [α-32P]UTP (5000 Ci/mmol, Perkin–Elmer). For Pol III-specific transcription, assays contained 500 ng of template DNA (pU6-shLuc), 30 μg of nuclear extract proteins, 12 mM HEPES-KOH (pH 7.9), 0.14 mM EDTA, 5 mM MgCl2, 60 mM KCl, 1 mM creatine phosphate, 3 mM DTT, 12% (v/v) glycerol, 0.4 mM each of ATP, GTP and CTP, 0.004 mM UTP and 0.1 μCi [α-32P]UTP (5000 Ci/mmol, Perkin–Elmer). Transcription reactions were carried out in the absence or presence of α-amanitin (200 μg/ml, Roche). After incubation for 60 min at 30°C, RNA was extracted and analyzed on non-denaturing 6% polyacrylamide gels.
Transfections
For the reporter assays, we transfected different human (HEK 293, HeLa) and mouse cell lines (HT22, HN9, Neuro-2a). Cells were plated onto 24-well plates at a density of 2 × 105 cells/well (HEK293 cells) or 8 × 104 cells/well (HT22 cells). HN9, Neuro-2a and HeLa cells were plated at different cell numbers. They where grown until they reached a confluency of 90–95% and then transfected. Each well contained 500 μl of the respective culture medium but without antibiotics/antimycotics. Cells were transfected either with Effectene (Qiagen) or Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol.
For analysis in western blots, HN9 cells were cultured and transfected in the same way as above but in 6-well format using 2 ml of medium per well. To achieve a high transfection efficiency, we additionally transfected mouse ES cells (IDG3.2 ES cells) via electroporation. Thereby, 2 × 106 cells were used in a volume of 800 μl phosphate-buffered saline (PBS) by applying an electrical field of 330 V to 4 mm cuvettes (Bio-Rad). Afterwards cells were replated into one 10 cm culture dish and the medium was replaced 24 h later.
Plasmids pGL3-Con and pRL-SV40 expressing firefly and Renilla luciferases, were purchased from Promega. pU6 empty vector and pU6-shLuc expressing shRNA targeting firefly luciferase (pSHAG and pSHAG-FF, respectively in original nomenclature) were kindly provided by Prof. G. Hannon (Cold Spring Harbor Laboratory, Cold Spring Harbor, NY). The total amount of plasmid DNA per well was kept equal and adjusted either with pBluescript (Stratagene) or pcDNA3 (Invitrogen) if needed. All transfections were done at least in duplicate.
Dual luciferase assay
Firefly and Renilla luciferase activities were measured using the Dual luciferase assay kit (Promega) according to the manufacturer's instructions. Briefly, 24–48 h after transfection, 100 μl of 1× passive lysis buffer (PLB) were dispensed per well on a 24-well plate. Culture plates were incubated for 15 min at room temperature, and 50 μl of the cell lysate were used to assess luciferase activities. The reconstituted 50× Stop&Glo substrate was diluted 1:100 with Stop &Glo buffer. To the cell lysate in the microtiter plate, 50 μl of LARII reagent were added (Microlite). Finally, 50 μl Stop&Glo reagent were applied to the lysates.
Western blot analysis
Western blot analysis was performed with protein extracts from lipid-mediated transfected HN9 and electroporated IDG3.2 ES cells. HN9 cells were collected 36–48 h after transfection. Cells were washed once with cold PBS, collected in 500 μl PBS using a cell scraper and transferred into an Eppendorf tube. Following centrifugation, cells were resuspended in Ampuwa water with protease inhibitors (Roche Mini). Cells from 3 wells of a 6-well plate were pooled in a total volume of 60 μl and sonicated. The lysates were cleared by centrifugation (5 min/16.000 × g/4°C) and the protein content was quantified by Bradford reagent (Bio-Rad).
IDG3.2 ES cells from each 10 cm plate were lysed at 48 h after transfection in 500 μl lysis buffer [2% SDS, 50 mM Tris (pH 6.8), 50 mM DTT, 10% glycerol, protease inhibitors (Roche Mini), 0.01% bromophenol blue]. Protein concentration was determined in lysis buffer without dye using the BCA assay kit (Pierce).
From both cell types, 20 μg of each protein lysate were separated by gel electrophoresis (SDS–PAGE) and transferred onto an immobilon polyvinylidene difluoride (PVDF) membrane (Millipore, Bedford, MA). Membranes were blocked with 5% non-fat dried milk powder (Nestlé, Vevey, Switzerland) in TBS/T (0.05% Tween-20; Bio-Rad) for 2 h at room temperature. The primary antibody recognizing ERK 1/2 as epitope (p44/42 MAP kinase, Thr202/Tyr204, Cell Signaling Technology, Beverly, MA) was diluted 1:1000 in TBS/T with 5% non-fat dried milk and incubated for 2 h at room temperature. Afterwards, the blots were washed two times for 10 min each with washing buffer (TBS/T) and then incubated with horseradish peroxidase (HRP)-conjugated anti-rabbit antibody (GE Healthcare, Buckinghamshire, UK). The secondary antibody was diluted 1:4000 in TBS/T 5% non-fat dried milk), incubated for 2 h at room temperature and unbound antibody was removed by washing as above. The immune complex formed was detected by ECL Plus (GE Healthcare, Buckinghamshire, UK) according to the manufacturer's protocol and the chemiluminescent signal was exposed to X-ray film (Fuji Photo Film, Tokyo, Japan). We performed a standard curve using defined steps of increased luminescence activity to assess the linear range of the used films. For further analysis, only films with optical densities in the estimated linear range of the film were used and scanned with a CanoScan 9900F. The band intensities were finally quantified using TINA Software (Raytest).
RESULTS
We present here a new vector suitable for expression of shRNAs. Dicer processes shRNAs to siRNAs, which serve as effector molecules in gene silencing (22). In order to be successfully recognized and processed by Dicer, shRNAs need to satisfy strict sequence and structure requirements. In particular, it is very important to fix precisely the transcription initiation and termination points, so that no nucleotide overhangs are present at either end of the resulting hairpin. The rules governing Pol I-dependent transcription have been extensively studied, making the design of such an exact transcript possible (18). Mouse Pol I promoter encompasses sequences downstream of the transcription start, which are, however, not required for efficient transcription. Only the presence of a purine nucleotide at position +1 appears to be indispensable (23). The transcription termination point is also precisely determined, being 15 bp upstream from the so called Sal box terminator (23). The Pol I minigene on pW contains a complete mouse Pol I promoter and several additional sequences between promoter and transcription termination point. The cloning strategy aimed at removing these additional sequences (Figure 1). After a two-step PCR procedure, we subcloned the Pol I promoter with termination sequences (Tx) into either pW or pE3SP backbone. The latter contains an enhancer sequence, which increases the efficiency of Pol I-dependent transcription (21). We called these vectors pW-bc and pE3SP-bc, respectively (Figure 2A). Between the promoter and the termination sequence Tx, these vectors contain KpnI and BbsI sites, which were used to insert a MCS, resulting in plasmids pW-MCS and pE3SP-MCS, respectively (Figure 2B). The two BbsI sites of the pE3SP-MCS were used for cloning of the short hairpin sequence targeting firefly luciferase. The sequence is the same as published by Paddison et al. (24). The construct was termed pE3SP-shLuc. Plasmid pE3SP-MCS was used as control in experiments with silencing firefly luciferase and Erk2 (Figures 2, 4 and 5; see below). The BbsI restriction enzyme offers the advantage that the cleavage site is positioned outside the recognition sequence of the nuclease; consequently, no strict sequence requirements are needed for the exact cleavage at the desired position. Moreover, BbsI cleavage generates non-palindromic cohesive ends, thus allowing directional cloning.
Next, we checked for the expression of the shLuc hairpin in an in vitro transcription assay using whole nuclear extracts. pE3SP-shLuc was used as a template, and 32P-labeled UTP was incorporated into the primary transcript. As expected, a fragment of 60 nt was detected, corresponding to the unprocessed hairpin precursor shLuc (Figure 3A), but not the mature siRNAs. Same results were obtained in an in vitro transcription assay using nuclear extracts, partially purified by DEAE ion exchange chromatography (data not shown). The lack of the mature siRNA in the nucleus is consistent with the notion that final processing to siRNA by Dicer takes place in the cytoplasm. As previously suggested (25,26), the Pol I promoter might contain cryptic recognition sites for other types of polymerases, in particular Pol II. However, in our case, transcription was not sensitive to treatment with α-amanitin, an inhibitor of both Pol II- and Pol III-dependent, but not of Pol I-dependent transcription. This observation strongly suggests that transcription from pE3SP-shLuc was indeed mediated by Pol I activity. In contrast, when the Pol III promoter-containing pU6-shLuc (Figure 3A and B) and 5S rRNA (Figure 3C) were used as templates, the transcription was completely inhibited by α-amanitin. Notably, two α-amanitin-sensitive signals were observed, when using pU6-shLuc as a template. The 60 nt transcript corresponded to the expected shLuc transcript, while the transcript of 150 nt indicates that initiation or/and termination of transcription have occurred at yet unidentified sites.
Figure 3.
In vitro transcription assay using plasmids pE3SP-shLuc and pU6-shLuc in nuclear extracts. Assays were done in absence or presence of α-amanitin, a specific inhibitor of Pol II- and Pol III-dependent transcription. (A) The expected band of 60 nt, corresponding to the expressed shRNA. (B) Additional fragment of 150 nt in length, resulting from pU6-shLuc (Pol III-dependent) transcription. (C) 5S rRNA transcript (Pol III-dependent), used as a positive control for assay, and inhibition by α-amanitin. Asterisk indicates an unspecific signal, inherent to the transcription from nuclear extract of FM3A cells.
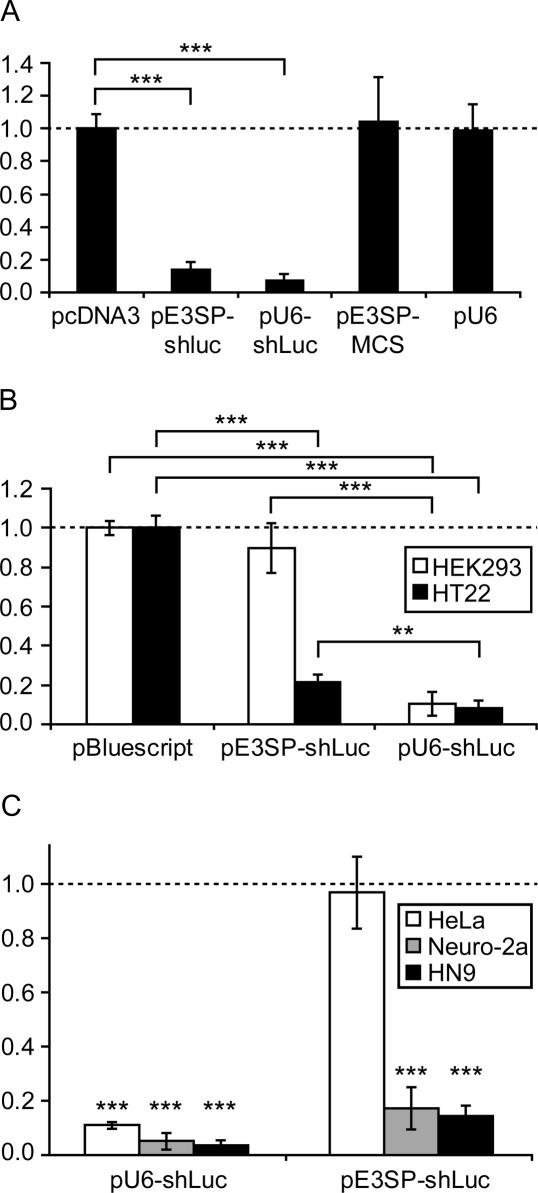
Then, functional efficiency of pE3SP-shLuc in mediating silencing was investigated. We co-transfected pE3SP-shLuc into the mouse hippocampal cell line HT22 with the vectors pGL3-Con expressing firefly luciferase (the target of shLuc) and pRL-SV40 expressing Renilla luciferase, which serves as an internal reference to normalize transfection efficiency. The pE3SP-shLuc construct was able to specifically suppress the activity of firefly luciferase by 86% (Figure 4A). This efficiency of suppression was comparable to the Pol III-driven vector pU6-shLuc (24). The effect was dose-dependent for both silencing constructs and reached its optimal efficiency at a silencer to reporter ratio of 10:1 (data not shown).
Figure 4.
Pol I-dependent shRNA induces species-specific gene silencing. (A) Twenty four-well plates of HT22 mouse hippocampal cells were transfected with 80 ng/well of plasmid that directs the expression of firefly (pGL3-Con, Promega), 8 ng/well of plasmid expressing Renilla (pRL-SV40, Promega) luciferase and 800 ng/well of the indicated DNA. Luciferase activities were assayed 48 h after transfection using the dual luciferase assay. Ratios of firefly to Renilla luciferase activities were normalized to a control transfected with vector pcDNA3, and the control ratio was set to 1. The average of two independent experiments is shown, and each experiment was performed in duplicate; error bars indicate SD. pE3SP-MCS and pU6 are empty vectors for corresponding shLuc-expressing plasmids pE3SP-shLuc and pU6-shLuc, respectively. ***P < 0.001 Statistical significance was evaluated using the Student's t-test. (B) Species specificity of Pol I-dependent gene silencing. Twenty four-well plates of HT22 mouse hippocampal cells or HEK293 human embryonic kidney cells were transfected and the luciferase expression was measured as in (A). Ratios of firefly to Renilla luciferase activities were normalized to a control transfected with pBluescript vector. The average of two independent experiments is shown, and each experiment was performed in duplicate; error bars indicate SD. ***P < 0.001, **P < 0.005. (C) Species specificity of Pol I-dependent gene silencing. Twenty four-well plates of HeLa human epithelial cells, Neuro-2a mouse neuroblastoma cells or HN9 mouse embryonic hippocampal cells were transfected, and luciferase expression was measured as in (A). Ratios of firefly to Renilla luciferase activities are expressed as a percentage of the control sample that was transfected with the accordant empty expression vector (pU6 for pU6-shLuc and pE3SP-MCS for pE3SP-shLuc). The average of two independent experiments is shown, and each experiment was performed in triplicate; error bars indicate SDs. Statistical analysis was done using the Student's t-test and asterisks indicate significant difference from the empty vector control. ***P < 0.001.
Unlike Pol II and Pol III, Pol I transcription exhibits a remarkable species specificity, e.g. between mouse and human (19). To test whether this species specificity is maintained in the Pol I-dependent silencing constructs, silencing efficiencies of pE3SP-shLuc and pU6-shLuc were compared in mouse HT22 and human HEK293 cells. Whereas human Pol III promoter-driven silencer pU6-shLuc was efficient in cell lines of both species, pE3SP-shLuc was only active in mouse, but completely silent in human cells (Figure 4B). In order to distinguish the cell line-specific effect from the species-specific one, we repeated this experiment in two additional mouse (HN9 and Neuro-2a), and one human (HeLa) cell lines. Again, the silencing was only efficient in mouse cells, and absent in the human cell line (Figure 4C). This observation is a strong indication that transcription relies indeed on Pol I-mediated mechanisms.
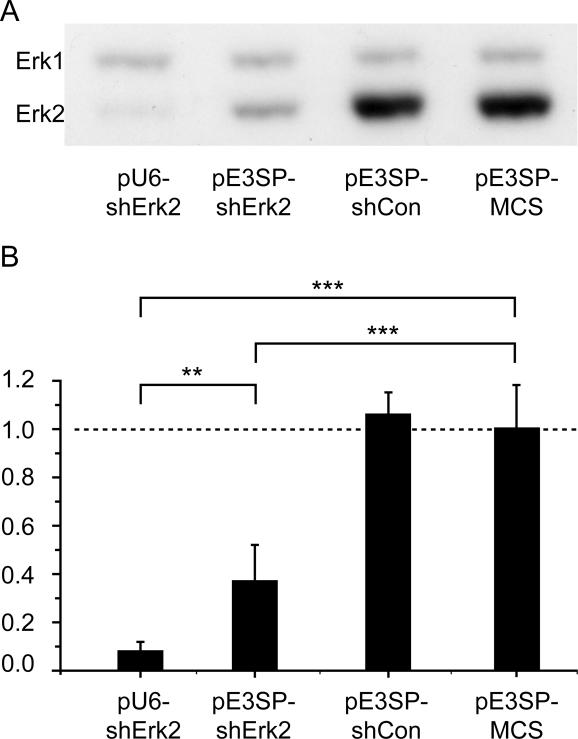
Finally, we tested our new expression cassette for the capacity to silence an endogenous gene. As a target, we took Erk2, a p42 MAP kinase. The hairpin sequence was already tested for the silencing activity in the Pol III promoter-driven vector (R. Kühn, unpublished data). We subcloned the shRNA, targeting Erk2, into the pE3SP-MCS vector backbone, and tested it for silencing activity in the IDG3.2 mouse ES cells and the HN9 mouse cell line (Figure 5). pE3SP-shLuc could significantly down-regulate the expression of Erk2, although the efficiency of silencing was lower as compared to Pol III-based U6-shErk2. Silencing was also highly specific, as two hairpins targeting unrelated sequences were unable to produce any effect on Erk2 expression (pE3SP-shCon in Figure 5). We could reproduce these findings in independent experiments with transfection of silencers in HN9 mouse cell line, where both Pol III and Pol I shRNAs showed similar effects in Erk2 down-regulation (data not shown).
Figure 5.
Pol I-dependent shRNA induces silencing of endogenous gene expression. (A) Western blot using 20 μg of protein extracts from electroporated IDG3.2 ES cells. A total of 50 μg of each of the plasmids pU6-shErk2, pE3SP-shErk2, pE3SP-shCon and pE3SP-MCS were transfected. The western blot shows the expected double-band for Erk1 (upper band) and Erk2 (lower band). Erk2 is specifically down-regulated in the samples from the transfected Pol III-driven (first lane) and Pol I-driven (second lane) RNAi expression vectors. Erk1 expression remains unaffected by the introduced RNAi and control vectors (lanes 3 and 4). (B) Quantification of band intensities in western blot shown in (A). The numerical values from the band intensities of the Erk2 signals were normalized to those of Erk1 (mean ± SD; Student's t-test: ***P < 0.001, **P < 0.01). There is a significant and strong down-regulation of Erk2 by shRNAs either derived from Pol III (first column) or Pol I promoters (second column). Unrelated shRNAs (third column) and empty expression vectors (fourth column) show no statistical effect on the expression of Erk2.
DISCUSSION
In this work, we generated a ribosomal minigene-based vector for expression of shRNA under the control of a mouse Pol I promoter. We showed the presence of the primary transcript of correct length expressed from this new vector, termed pE3SP-shLuc. We also demonstrated the functionality of our system in efficient silencing of firefly luciferase gene in mouse but not in human cells by using the pE3SP-shLuc. Finally, we proved the efficacy and specificity of the expression vector in silencing the endogenous Erk2 expression. The formation of the primary transcript is not sensitive to inhibition by the Pol II- and Pol III-specific toxin α-amanitin. This proves that the expression is initiated by a Pol I-specific promoter, and not by cryptic promoters for class II and class III RNA polymerases, whose activity might interfere with Pol I activity, as suggested previously (25,26).
Interestingly, we found both the expected and longer primary transcripts, when a Pol III-dependent vector was used as a template in in vitro transcription assay. This fact might suggest a read-through from the plasmid, probably due to leaky transcription termination in this vector. It was noted that the termination of transcription of Pol III relies strongly on the exact sequence of the termination signal and the proximity of the promoter (27,28). As the shRNA insert is very short, it might lead to interference between the enzymatic machineries responsible for transcription initiation and termination. It would be interesting to investigate whether this longer transcript is exported to the cytoplasm and can serve as a template for further processing by Dicer, resulting in the generation of functional siRNAs.
The species specificity of Pol I-dependent transcription provides an important advantage in terms of experimental biosafety, as compared to other types of vectors. RNAi-mediated down-regulation is frequently used to systematically reveal the function of unknown genes, often using viral vectors, such as lentivirus (29–32). Currently used lentiviral vectors are designed to inactivate themselves once integrated into the genome. However, they still preserve the capacity to infect cells, including those of the human experimenter. The use of the mouse-specific Pol I promoter, which is silent in human cells, represents a considerable advantage. The next logical step in the development of Pol I-based expression vectors will be the generation of the Tet-inducible cassette to allow a regulated knock-down.
Pol III-based vectors are now extensively used to express shRNAs in numerous systems (33). However, an alternative method to express short RNA molecules may be required for particular experiments. A need to screen or modify Pol III promoters in order to obtain a strong and reliable inhibition in a particular cell type or organ has been reported (34,35). Occasionally, irreversible silencing of Pol III-dependent transcription was observed in stable expression systems. Thus, it would be desirable to systematically compare different Pol III- with Pol I-dependent vectors regarding efficiency, stability and toxicity issues.
To further broaden the scope of Pol I-driven expression vectors, we constructed other minimal ribosomal minigene vectors containing a MCS. Potentially, they can be used to express other types of functional RNA molecules, such as snoRNAs or viral RNAs (23). Finally, it would be desirable to generate a reporter vector, expressing for example luciferase downstream of an IRES signal for cap-independent translation. Up to now, the attempts to create such a reporter largely failed because of above-mentioned reservations concerning the presence of cryptic Pol II recognition sites in ribosomal minigenes. Only one publication reported a construction of such a vector, based on human Pol I promoter (36). However, our results strongly point to the specificity of Pol I-dependent transcription from our vectors.
In conclusion, we believe that our Pol I-driven expression vectors will be a useful addition to the palette of RNAi-based tools for silencing gene function.
Acknowledgments
The authors wish to thank the VolkswagenStiftung (Az.: I/78 767, Az.: I/77 114) for the generous support of this work, Jérome Cavaillé and Greg Hannon for providing plasmids used in this study, Barbara Wölfel and Bettina Dörr for the excellent technical assistance, Jan Deussing for mediating the people and meetings related to this work, Florian Riese for critical reading of the manuscript and Stefano Brenz Verca for sharing his knowledge and experience in numerous discussions throughout the course of this work. Funding to pay the Open Access publication charges for this article was provided by the VolkswagenStiftung.
Conflict of interest statement. None declared.
REFERENCES
- 1.Baulcombe D. RNA silencing. Trends Biochem. Sci. 2005;30:290–293. doi: 10.1016/j.tibs.2005.04.012. [DOI] [PubMed] [Google Scholar]
- 2.Hannon G.J., Rossi J.J. Unlocking the potential of the human genome with RNA interference. Nature. 2004;431:371–378. doi: 10.1038/nature02870. [DOI] [PubMed] [Google Scholar]
- 3.Mello C.C., Conte D., Jr Revealing the world of RNA interference. Nature. 2004;431:338–342. doi: 10.1038/nature02872. [DOI] [PubMed] [Google Scholar]
- 4.Zamore P.D., Tuschl T., Sharp P.A., Bartel D.P. RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell. 2000;101:25–33. doi: 10.1016/S0092-8674(00)80620-0. [DOI] [PubMed] [Google Scholar]
- 5.Elbashir S.M., Lendeckel W., Tuschl T. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 2001;15:188–200. doi: 10.1101/gad.862301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hamilton A.J., Baulcombe D.C. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science. 1999;286:950–952. doi: 10.1126/science.286.5441.950. [DOI] [PubMed] [Google Scholar]
- 7.Parrish S., Fleenor J., Xu S., Mello C., Fire A. Functional anatomy of a dsRNA trigger: differential requirement for the two trigger strands in RNA interference. Mol. Cell. 2000;6:1077–1087. doi: 10.1016/s1097-2765(00)00106-4. [DOI] [PubMed] [Google Scholar]
- 8.Hutvagner G., Zamore P.D. RNAi: nature abhors a double-strand. Curr. Opin. Genet. Dev. 2002;12:225–232. doi: 10.1016/s0959-437x(02)00290-3. [DOI] [PubMed] [Google Scholar]
- 9.Caplen N.J., Parrish S., Imani F., Fire A., Morgan R.A. Specific inhibition of gene expression by small double-stranded RNAs in invertebrate and vertebrate systems. Proc. Natl Acad. Sci.USA. 2001;98:9742–9747. doi: 10.1073/pnas.171251798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elbashir S.M., Harborth J., Lendeckel W., Yalcin A., Weber K., Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 11.Verma N.K., Dey C.S. RNA-mediated gene silencing: mechanisms and its therapeutic applications. J. Clin. Pharm. Ther. 2004;29:395–404. doi: 10.1111/j.1365-2710.2004.00579.x. [DOI] [PubMed] [Google Scholar]
- 12.O'Neil N.J., Martin R.L., Tomlinson M.L., Jones M.R., Coulson A., Kuwabara P.E. RNA-mediated interference as a tool for identifying drug targets. Am. J. Pharmacogenomics. 2001;1:45–53. doi: 10.2165/00129785-200101010-00006. [DOI] [PubMed] [Google Scholar]
- 13.Brummelkamp T.R., Bernards R., Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science. 2002;296:550–553. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- 14.Lee N.S., Dohjima T., Bauer G., Li H., Li M.J., Ehsani A., Salvaterra P., Rossi J. Expression of small interfering RNAs targeted against HIV-1 rev transcripts in human cells. Nat. Biotechnol. 2002;20:500–505. doi: 10.1038/nbt0502-500. [DOI] [PubMed] [Google Scholar]
- 15.Paul C.P., Good P.D., Winer I., Engelke D.R. Effective expression of small interfering RNA in human cells. Nat. Biotechnol. 2002;20:505–508. doi: 10.1038/nbt0502-505. [DOI] [PubMed] [Google Scholar]
- 16.Miyagishi M., Taira K. U6 promoter-driven siRNAs with four uridine 3′ overhangs efficiently suppress targeted gene expression in mammalian cells. Nat. Biotechnol. 2002;20:497–500. doi: 10.1038/nbt0502-497. [DOI] [PubMed] [Google Scholar]
- 17.Zhou H., Xia X.G., Xu Z. An RNA polymerase II construct synthesizes short-hairpin RNA with a quantitative indicator and mediates highly efficient RNAi. Nucleic Acids Res. 2005;33:e62. doi: 10.1093/nar/gni061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grummt I. Regulation of mammalian ribosomal gene transcription by RNA polymerase I. Prog. Nucleic Acid Res. Mol. Biol. 1999;62:109–154. doi: 10.1016/s0079-6603(08)60506-1. [DOI] [PubMed] [Google Scholar]
- 19.Heix J., Grummt I. Species specificity of transcription by RNA polymerase I. Curr. Opin. Genet. Dev. 1995;5:652–656. doi: 10.1016/0959-437x(95)80035-2. [DOI] [PubMed] [Google Scholar]
- 20.Hadjiolova K.V., Normann A., Cavaille J., Soupene E., Mazan S., Hadjiolov A.A., Bachellerie J.P. Processing of truncated mouse or human rRNA transcribed from ribosomal minigenes transfected into mouse cells. Mol. Cell. Biol. 1994;14:4044–4056. doi: 10.1128/mcb.14.6.4044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuhn A., Deppert U., Grummt I. A 140-base pair repetitive sequence element in the mouse rRNA gene spacer enhances transcription by RNA polymerase I in a cell-free system. Proc. Natl Acad. Sci. USA. 1990;87:7527–7531. doi: 10.1073/pnas.87.19.7527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bernstein E., Caudy A.A., Hammond S.M., Hannon G.J. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 23.Zobel A., Neumann G., Hobom G. RNA polymerase I catalysed transcription of insert viral cDNA. Nucleic Acids Res. 1993;21:3607–3614. doi: 10.1093/nar/21.16.3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paddison P.J., Caudy A.A., Bernstein E., Hannon G.J., Conklin D.S. Short hairpin RNAs (shRNAs) induce sequence-specific silencing in mammalian cells. Genes Dev. 2002;16:948–958. doi: 10.1101/gad.981002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smale S.T., Tjian R. Transcription of herpes simplex virus tk sequences under the control of wild-type and mutant human RNA polymerase I promoters. Mol. Cell. Biol. 1985;5:352–362. doi: 10.1128/mcb.5.2.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lopata M.A., Cleveland D.W., Sollner-Webb B. RNA polymerase specificity of mRNA production and enhancer action. Proc. Natl Acad. Sci. USA. 1986;83:6677–6681. doi: 10.1073/pnas.83.18.6677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Myslinski E., Ame J.C., Krol A., Carbon P. An unusually compact external promoter for RNA polymerase III transcription of the human H1RNA gene. Nucleic Acids Res. 2001;29:2502–2509. doi: 10.1093/nar/29.12.2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gunnery S., Ma Y., Mathews M.B. Termination sequence requirements vary among genes transcribed by RNA polymerase III. J. Mol. Biol. 1999;286:745–757. doi: 10.1006/jmbi.1998.2518. [DOI] [PubMed] [Google Scholar]
- 29.Li M., Rossi J.J. Lentiviral vector delivery of siRNA and shRNA encoding genes into cultured and primary hematopoietic cells. Meth. Mol. Biol. 2005;309:261–272. doi: 10.1385/1-59259-935-4:261. [DOI] [PubMed] [Google Scholar]
- 30.Raoul C., Abbas-Terki T., Bensadoun J.C., Guillot S., Haase G., Szulc J., Henderson C.E., Aebischer P. Lentiviral-mediated silencing of SOD1 through RNA interference retards disease onset and progression in a mouse model of ALS. Nature Med. 2005;11:423–428. doi: 10.1038/nm1207. [DOI] [PubMed] [Google Scholar]
- 31.Taulli R., Accornero P., Follenzi A., Mangano T., Morotti A., Scuoppo C., Forni P.E., Bersani F., Crepaldi T., Chiarle R., et al. RNAi technology and lentiviral delivery as a powerful tool to suppress Tpr-Met-mediated tumorigenesis. Cancer Gene Ther. 2005;12:456–463. doi: 10.1038/sj.cgt.7700815. [DOI] [PubMed] [Google Scholar]
- 32.Fish R.J., Kruithof E.K. Short-term cytotoxic effects and long-term instability of RNAi delivered using lentiviral vectors. BMC Mol. Biol. 2004;5:9. doi: 10.1186/1471-2199-5-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miyagishi M., Taira K. RNAi expression vectors in mammalian cells. Meth. Mol. Biol. 2004;252:483–491. doi: 10.1385/1-59259-746-7:483. [DOI] [PubMed] [Google Scholar]
- 34.Paul C.P. Subcellular distribution of small interfering RNA: directed delivery through RNA polymerase III expression cassettes and localization by in situ hybridization. Meth. Enzymol. 2005;392:125–145. doi: 10.1016/S0076-6879(04)92008-3. [DOI] [PubMed] [Google Scholar]
- 35.Boden D., Pusch O., Lee F., Tucker L., Shank P.R., Ramratnam B. Promoter choice affects the potency of HIV-1 specific RNA interference. Nucleic Acids Res. 2003;31:5033–5038. doi: 10.1093/nar/gkg704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Palmer T.D., Miller A.D., Reeder R.H., McStay B. Efficient expression of a protein coding gene under the control of an RNA polymerase I promoter. Nucleic Acids Res. 1993;21:3451–3457. doi: 10.1093/nar/21.15.3451. [DOI] [PMC free article] [PubMed] [Google Scholar]