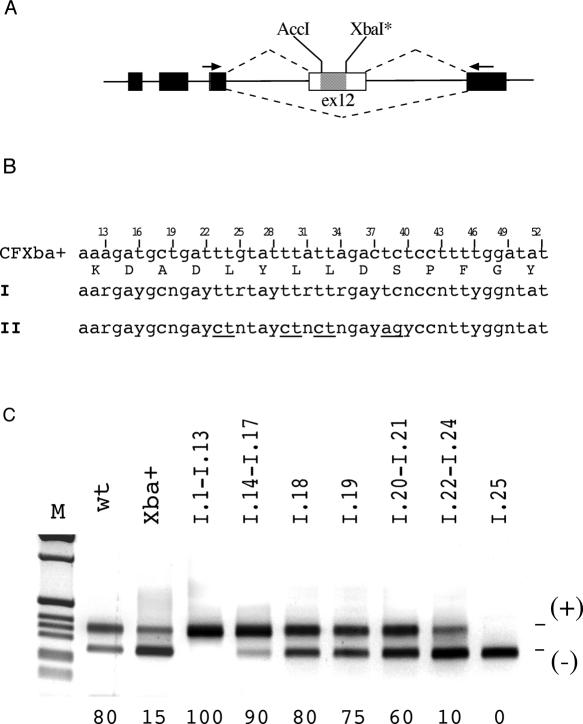
Figure 1.
Combined effect of multiple random synonymous substitutions on CFTR exon 12 splicing pattern. (A) Schematic representation of the CFTR hybrid minigene used in transient transfection assay showing the location of the two restriction sites in exon 12 that were used for cloning the I and II degenerated oligonucleotides. Exonic and intronic sequences are shown as boxes and lines, respectively, the hatched box corresponds to the substituted sequence and the arrows show the specific oligonucleotides used in RT–PCR analysis. The two alternative splicing possibilities are indicated as dotted lines. The XbaI site was created by site-directed mutagenesis and corresponds to 52T synonymous substitution. (B) Nucleotide composition of part of the human CFTR exon 12 between the AccI and XbaI sites. Third nucleotide positions are numbered according to their location in the exon. The two 14-degenerated codons oligonucleotides I and II (II differs to I at conserved Leu and Ser codons, which are underlined) are indicated. These oligomers were cloned in the CFTR exon 12 to create random changes at synonymous sites between the exonic positions 13 and 52. (C) Effect of multiple random synonymous substitutions on splicing efficiency. Representative samples of the CFTR exon 12 minigenes derived from the degenerated oligonucleotides I are shown and their sequences are represented in Figure 2. Minigenes were transfected in Hep3B cells and analyzed for splicing efficiency. The percentages of exon inclusion are reported at the bottom of each lane and are the mean of three independent experiments done in duplicate. The effect of the multiple random synonymous substitutions is compared with the WT and the CFXba+ minigenes. The exon 12 inclusion and skipping forms are indicated.