Figure 3.
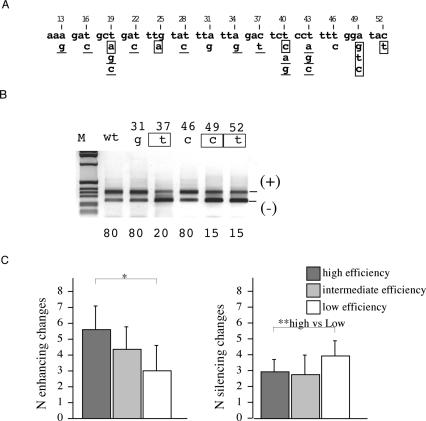
The effect of multiple synonymous substitutions on splicing correlates with the sum of the effect of each single substitution. (A) Nucleotide composition of part of the human CFTR exon 12 between the AccI and XbaI sites. Third nucleotide positions are numbered according to their location in the exon. The 22 possible single synonymous changes are shown. According to their individual effect on splicing in WT context, the synonymous substitutions were classified as positive, (mutations that induce exon inclusion, underlined) negative (exon skipping, boxed ) or neutral (no effect in black). (B) Representative RT–PCR products from transfection experiments showing the effect of single synonymous substitutions. The 31g, 46c and 49c mutations, not tested previously (28), are compared with WT and other selected mutants. The exon 12 inclusion and skipping forms are indicated. The percentages of exon inclusion are reported at the bottom of each lane and are the mean of three independent experiments done in duplicate. (C) Correlation between splicing efficiency of single and multiple synonymous substitutions. The clones shown in Figure 2 with random multiple substitutions originated from the I oligonucleotides were grouped according to their splicing efficiency: high, intermediate and low corresponding to 100, 99–50 and 50–0% of splicing, respectively. For each group the number of positive or negative synonymous substitutions were calculated (y axis) and analyzed for statistical significance (*, P < 0.05 between the three different groups), (**, P < 0.05 between the high ad low efficiency groups).