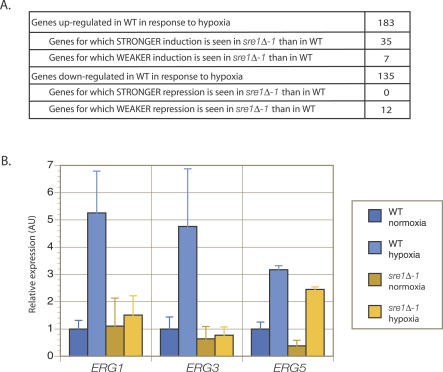
Figure 5. Microarray Hybridization and RT-QPCR Studies Demonstrate That a Subset of Genes Involved in the Hypoxia Response Require Sre1 for Their Regulation.
(A) Summary of profiling transcriptional response to hypoxia in sre1Δ and wild-type (WT) cells. cDNA from wild-type cells cultivated in normoxic conditions was hybridized against cDNA from wild-type cells exposed to hypoxic conditions on arrays containing 6,846 features from the genome of C. neoformans. Similarly, cDNA from sre1Δ-1 grown in normoxia was hybridized against cDNA from sre1Δ-1 grown in hypoxia. Statistical analysis of the resulting arrays was conducted using the software SAM. The transcriptional response to hypoxia in wild-type was identified as the set of genes with statistically significant changes in gene expression as determined by SAM. A cutoff of 2-fold or greater changes in gene expression was imposed upon this gene set to determine the number of genes up- and down-regulated in wild-type in response to hypoxia. The arrays hybridized with cDNA from sre1Δ-1 were compared to those hybridized with wild-type cDNA to determine which hypoxia-induced transcriptional changes were significantly different between sre1Δ-1 and wild-type. The resulting gene set is summarized above, where relative degree of induction/repression in sre1Δ-1 and wild-type were compared by dividing fold-change in expression in sre1Δ-1 with fold-change in expression in wild-type.
(B) RT-QPCR. cDNA from sre1Δ-1 and wild-type cells grown in normoxia and hypoxia were amplified using primers against ERG1, ERG3, ERG5, and ACT1. Values obtained for ERG1, ERG3, and ERG5 were normalized against ACT1 for each sample to give relative expression, and then expression for the three genes were normalized to their expression in wild-type in normoxic conditions. Error bars represent standard deviation across four samples.