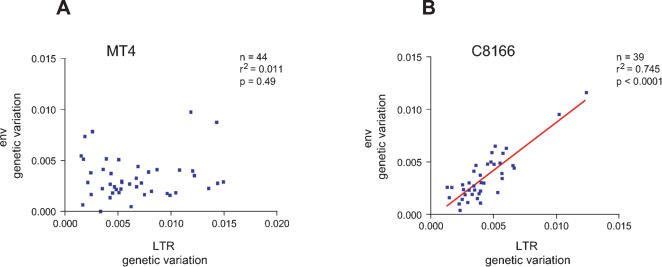
Figure 2. Genetic variation in the Long Terminal Repeat (LTR) and envelope region (Env) of MT4 and C8166 cultured viruses.
For each culture, ten clones of both the LTR and Env region were sequenced and this was repeated for five points in time. Thus, each data point corresponds to the amount of genetic variation within a single serial passage line at a certain point in time. Genetic variation (Kimura 2 parameter) is plotted separately for the two cell-lines. A) No correlation in genetic variation in MT4 evolved viruses was observed (n = 44, r2 = 0.011, p = 0.49). B) There is a strong linear correlation in genetic variation between the LTR and Env in C8166 cultured viruses (n = 39, r2 = 0.745, p<0.0001). Even if the two outliers are taken out, the strong correlation remains (n = 37, r2 = 0.475, p<0.0001).