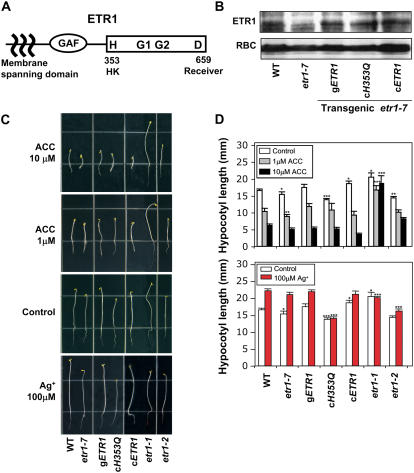
Figure 1.
Structure and functions of ETR1 HK activity for plant growth promotion in the dark. A, The diagram depicts putative functional domains of ETR1. B, Expression of ETR1 proteins was detected by protein-blot analysis using anti-ETR1 antibody (Santa Cruz Biotech). Transgenic lines are etr1-7 plants transformed with the wild-type gETR1, the cDNA ETR1 gene containing a H353Q mutation (cH353Q), and the wild-type cETR1. For comparison, proteins of wild type (ecotype Columbia) and etr1-7 (etr1 null) were analyzed together. To detect the membrane protein ETR1, plants (14 d) were ground in 50 mm HEPES-KOH buffer, pH 7.6, containing 2 mm dithiothreitol, 2 mm EDTA, 10 mm β-glycerophosphate, 20% (v/v) glycerol, 1× complete protease inhibitor mix (Roche), and 1% (v/v) Triton X-100, and incubated on ice for 30 min. After removing tissue debris, protein loading buffer was added. The extracts were incubated at 65°C for 15 min and then 37°C for 30 min before running on 10% (w/v) SDS-polyacrylamide gels. The Rubisco (RBC) was shown as a protein loading control (Yanagisawa et al., 2003). C, Phenotypes of 4-d-old seedling growth in the dark. Seedlings were grown under saturating dosage of ACC (10 μm), subsaturating dosage of ACC (1 μm), ambient/endogenous ethylene (control), and ethylene-free (100 μm Ag+) conditions. For comparison, wild-type, etr1-7 (etr1 null), etr1-1 (strong ethylene-insensitive), and etr1-2 (weak ethylene-insensitive) seedlings were grown together with the transgenic etr1-7 lines. D, Quantitative analysis of hypocotyl length of 4-d-old etiolated seedlings. Values are means with sd, n = 30. Asterisks over bars indicate differences between wild type and mutant with statistical significance at *P < 0.05, **P < 0.01, and ***P < 0.001 (t test). [See online article for color version of this figure.]