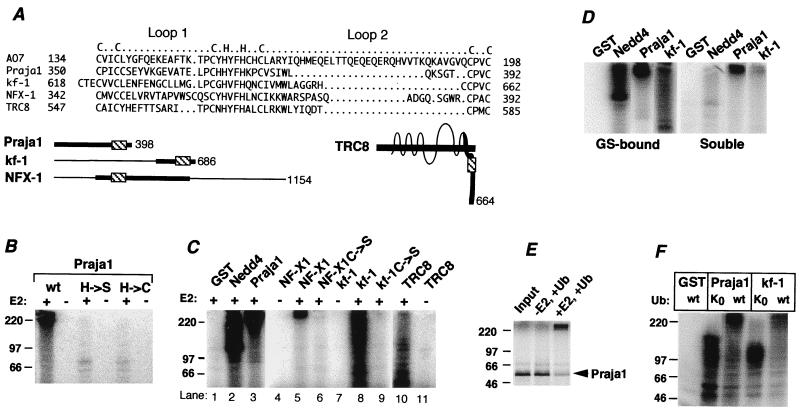
Figure 4.
Evaluation of four RING-H2 proteins. (A) Alignment of RING-H2 regions of proteins. Loops and predicted coordination sites are indicated above. kf-1 has two possible alignments beginning with either C618 or C621; the alignment shown begins with C621. Full-length forms of proteins are schematized below. Heavy lines indicate regions fused to GST (Praja1 is full length), and RING finger regions are indicated (▧). TRC8 is a multispanning membrane protein. (B) GST-Praja1 and mutants of the fifth putative coordination site to either Ser (H → S) or Cys (H → C) were evaluated for ubiquitination. (C) GST fusions of the indicated proteins were evaluated as described for B. The kf-1, NF-X1, and TRC8 fusions included amino acids 538–686, 207–645, and 477–664, respectively. The C → S mutation of kf-1 was at amino acid 621; NF-X1 C → S was a double mutation at C342 and C345. Because of difficulty expressing GST-NF-X1, only 5 pmol was used, and 20 pmol of NF-X1 C → S was used. (D) Soluble material was removed from ubiquitination reactions carried out as described for C, and GS beads were washed extensively. Both fractions were resolved by SDS/PAGE. (E) 35S-labeled Praja1 was resolved directly on SDS/PAGE (Input) or subjected to a ubiquitination reaction in E1-containing reaction buffer with or without UbcH5B. (F) GS-bound proteins were evaluated for ubiquitination with either wild-type Ub or UbK0. After Western transfer, membranes were immunoblotted with anti-Ub. GST-Praja1 and GST-kf-1 migrate at approximately 85 kDa and 50 kDa, respectively.