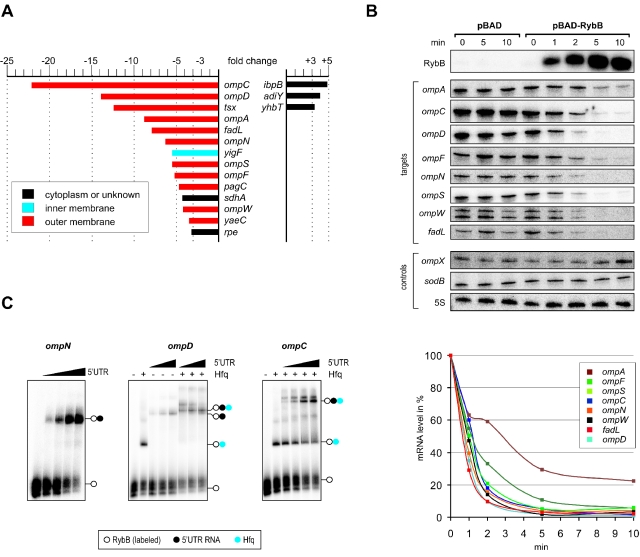
Fig. 3.
RybB targets a large set of mRNAs that encode OMPs. A. Fold changes of mRNA levels after RybB pulse overexpression. Salmonella carrying expression plasmid pBAD-RybB were cultured to an OD600 of 1.5, and RybB expression was induced with l-arabinose for 10 min. Total RNA was extracted and probed on Salmonella SALSA whole-genome microarrays, and data were analysed as described (Nagy et al., 2006). The fold changes given here are normalized to the mRNA expression changes of induced cells that carried a pBAD control vector, to correct for the effects of l-arabinose on global mRNA expression. The chart shows all mRNAs whose levels changed by > threefold, after statistical filtering (see Supplementary material for the entire data set). Bar colours indicate the predicted cellular localization of the encoded proteins. B. Northern blot validation of RybB-induced target mRNA decay. Salmonella carrying either the control pBAD vector or the pBAD-RybB expression vector were arabinose-induced at an OD of 1.5, and total RNA extracted at the time-points indicated above the panels. Northern hybridization with gene-specific probes (indicated to the left) confirmed rapid induction of RybB expression (upper panel), and a concomitant drop in the steady-state levels of eight target mRNAs (ompA/C/D/F/N/S/W, and fadL) in pBAD-RybB cells. The ompX and sodB mRNAs, whose expression did not change in the aforementioned microarray experiments, were probed as controls (panels below). Probing for 5S rRNA confirmed equal RNA loading (lower panel). A quantification of the blot signals obtained for the eight RybB target mRNAs in pBAD-RybB cells is given below. For each mRNA, the signal obtained at the 0 min time-point (prior to induction) was set to 100%. RybB expression reduces the half-life of these targets (except ompA) to ∼1 min. C. Gel-mobility shift assays show that RybB binds to RNA fragments derived from 5′ UTRs of three target mRNAs in vitro. RybB (32P-labelled; 5 nM final concentrations) binding assays with 5′ UTR RNAs were performed in the presence of a 1000-fold excess of yeast tRNA for 10 min at 37°C, followed by electrophoresis on a native gel, of which autoradiographs are shown. (Left panel) RybB was incubated with increasing concentrations of an unlabelled RNA fragment derived from the ompN 5′ UTR (from left to right: 0, 15, 30, 60, 120 nM). The positions of RybB (open circle) or the RybB/ompN complex (filled circle) are indicated to the right. (Middle panel) Gel mobility shifts with an unlabelled RNA derived from the ompD 5′ UTR (final concentration from left to right: 0, 0, 125, 250, 500, 125, 250, 500 nM). Here, Hfq was added to the individual reactions where indicated (+) at a final concentration of 30 nM; a blue circle indicates Hfq-specific mobility changes. (Right panel) Complex formation of RybB with an unlabelled RNA derived from the ompC 5′ UTR (final concentrations from left to right: 0, 0, 12.5, 25, 50, 100 nM). Addition of Hfq and complex formation is indicated as in the other two panels.