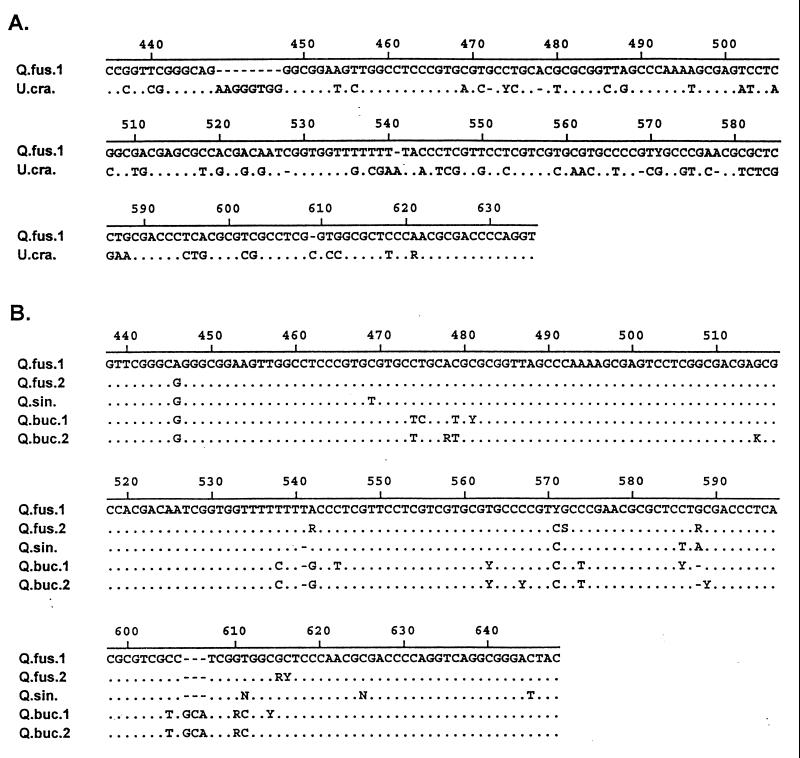
Figure 1.
Alignments of reference ITS sequences illustrating the ability to distinguish between genera (A) and species within a genus (B). (A and B) For both alignments, the top sequence is used as a reference for the sequences below it. Agreement with the reference sequence is indicated by a period, and differences are indicated by a nucleotide symbol. Gaps introduced to improve the alignment are indicated by dashes. Standard International Union of Biochemistry nucleotide symbols are used: A = adenosine, G = guanosine, C = cytidine, T = thymidine, R = purine (A or G), Y = pyrimidine (C or T), S = G or C, W = A or T, K = G or T, M = A or C, and N = any nucleotide (unreadable). All polymorphisms were confirmed on both strands. (A) Alignment of a portion of the ITS sequences for Q. fusiformis and U. crassifolia, the two most closely related genera in the reference database. All other intergeneric alignments showed even less similarity. (B) Alignment of three oak species: Q. fusiformis, Q. sinuata, and Quercus buckleyi. Note that there are two representatives for Q. fusiformis and Q. buckleyi. Although there are fewer diagnostic characteristics within the oaks, they can be distinguished readily by consistent differences across the ITS region (Q. fusiformis and Q. sinuata are the two most difficult species we distinguish). Vegetation surveys at the surface of each cave also constrain the species potentially found in each cave (and act as a check on the ITS results).