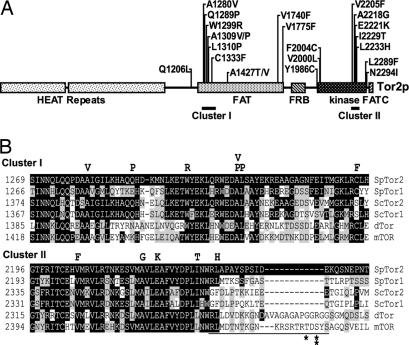
Fig. 2.
Location of activated mutations in Tor2p and conservation of residues. (A) All identified activating mutations are indicated above the linear representation of Tor2p. Clusters I and II are indicated. (B) TOR protein sequences from Schizosaccharomyces pombe (SpTor1 and SpTor2), S. cerevisiae (ScTor1 and ScTor2), Drosophila (dTOR), and human (mTOR) were aligned with MegAlign. Residues identical or similar to SpTor2 are shaded black or gray, respectively. Mutations found in SpTor2 are indicated above the alignment. The repressor domain region of mTOR is indicated by a line under the alignment. Single and double asterisks indicate the AMPK and S6K phosphorylation sites, respectively.