Fig. 1.
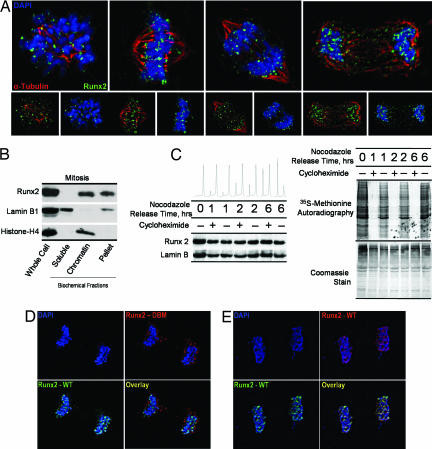
Runx2 is stable and associated with chromosomes during mitosis. (A) Asynchronously growing Saos-2 cells were fixed and stained for DNA by using DAPI and for Runx2 by using a rabbit polyclonal antibody. Mitotic cells were identified by chromosome morphology. High-resolution images obtained by three-dimensional deconvolution algorithms reveal that Runx2 (green) is localized in mitotic chromosomes. A subset of Runx2 colocalizes with the microtubules, labeled by α-tubulin staining (red). (B) Localization of Runx2 by biochemical fractionation using standard techniques to generate soluble, chromatin-associated, and insoluble protein fractions compared with whole-cell protein levels. Each fraction was analyzed by Western blotting using antibodies against Runx2, as well as lamin B1 and histone H4 as controls. (C) Stability of Runx2 protein in mitosis. Saos cells were arrested at the G2/M boundary by nocodazole treatment and released through mitosis into G1 in the presence or absence of the protein translation inhibitor cycloheximide (50 μg/ml). At the indicated times, protein synthesis was assayed by pulse labeling with [35S]methionine. In parallel, protein samples were isolated for Western blot analysis. (D and E) Colocalization studies of wild-type Runx2 and a DNA-binding mutant Runx2. HeLa cells were cotransfected with wild-type Runx2 and the R182Q mutant Runx2 (GenBank accession no. D14637), which were N-terminally tagged with HA and Xpress epitopes, respectively. In situ immunofluorescence microscopy was performed with DNA staining by DAPI and indirect immunolabeling with antibodies directed against HA and Xpress epitopes with appropriate secondary antibodies. Mitotic cells were identified by DNA morphology. Control colocalization experiments were performed by using HA- and Xpress-tagged wild-type proteins.