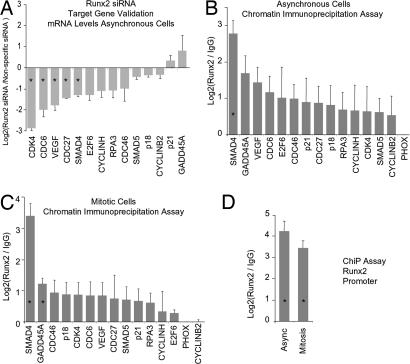
Fig. 3.
Target gene validation. (A) Fourteen putative Runx2 target genes identified in a primary screen (see Fig. 2) were tested for responsiveness to depletion of Runx2 by RNA interference. Independent siRNA experiments were analyzed in duplicate by RT-qPCR with primer sets for each of the target genes (SI Data Set 1). Expression data are normalized and displayed as the log2 difference between Runx2 and nonspecific siRNAs. Error bars reflect SEM. (B) Runx2 target genes were validated by ChIP in two independent experiments (SI Data Set 1). Samples were quantified by qPCR relative to input and normalized to nonspecific immunoprecipitation of the PHOX gene promoter. Values represent the log2 difference between Runx2-specific and control IgG signals. (C) Interaction of Runx2 with its novel target genes during mitosis was assessed by ChIP assays. Mitotic cells were isolated by nocodazole synchronization and mitotic shakeoff in two independent experiments, and samples were assayed in duplicate by qPCR. Data analysis is described in B. (D) Interaction of Runx2 with its promoter was assessed by ChIP on asynchronous and mitotic cells. Asterisks indicate statistical significance at the 0.05 level based on a t test.