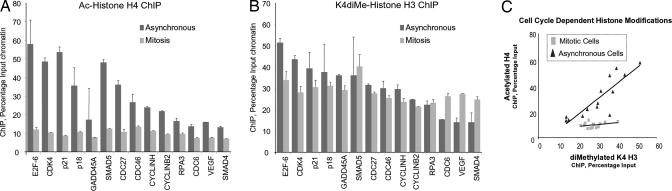
Fig. 4.
Runx2 is associated with epigenetically modified target genes in mitosis. Histone modifications at the 14 target genes were assayed by ChIP analyses of asynchronous (dark gray bars) and pure mitotic cells (light gray bars) synchronized as described in Fig. 3. Duplicate samples were analyzed by qPCR, quantified as a percentage of input, and normalized for comparison with subsequent functional experiments in Fig. 5. (A) Histone H4 acetylation and (B) histone H3–K4 dimethylation of gene promoters. (C) Scatterplot of H4 acetylation (ordinate) versus H3–K4 dimethylation (abscissa) for all 14 genes in asynchronous cells (black triangles) and mitotic cells (gray squares) is depicted. A least-squares regression line is shown for each population.