Fig. 5.
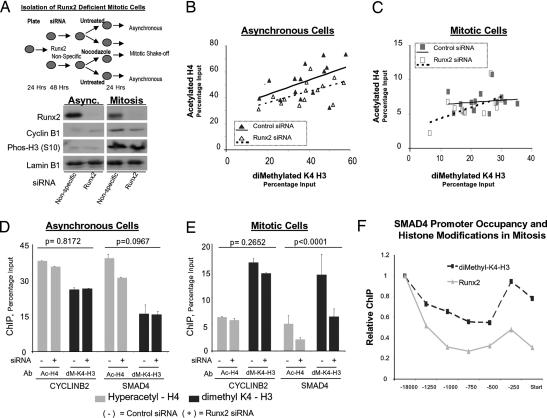
Runx2 affects mitotic histone modifications at target gene promoters. The effects of Runx2 on promoter histone modifications were assessed by combining siRNA gene knockdown with mitotic cell synchronization. (A) Experimental strategy to obtain Runx2-depleted mitotic cells. Histone modification levels at target gene promoters in mitotic and asynchronous cells were assayed by ChIP and analyzed by qPCR. Protein was extracted from parallel plates to validate Runx2 knockdown. Cyclin B1 levels and histone H3 (S10) phosphorylation status serve as markers of mitosis and lamin B1 as a loading control. Efficiency of siRNA transfection (>90%) was determined in parallel (data not shown). (B and C) Levels of hyperacetylated histone H4 and dimethylated histone H3 (K4) in control and Runx2 siRNA-treated cells were determined in two ChIP assays, each analyzed in duplicate by qPCR. Scatterplot of H4 acetylation versus H3 K4-dimethylation is shown for all 14 target genes in asynchronous (B) and mitotic cells (C) treated with control (black symbols) or Runx2 siRNA (open symbols). A least-squares regression line is shown for each population: control (solid line) or Runx2 siRNA (broken line). (D and E) A mixed-model ANOVA was used to assess significance of Runx2 siRNA effects for all target genes. Multiple pairwise comparisons (Tukey's HSD) were evaluated to determine which effects differ at a 0.05 level and to establish P values; error bars are SE (n = 4). Plots show ChIP results for SMAD4 and CYCLINB2 in Runx2 and control siRNA-treated asynchronous (D) and mitotic cells (E). (F) Binding of Runx2 across the SMAD4 promoter was compared with dimethyl-K4 histone H3 modifications. Primer sets encompassed positions in the proximal and distal SMAD4 promoter, as well as the transcription start site (see SI Data Set 1). Within-group (Runx2 or K4–H3 dimethylated) ratios as calculated in D and E are plotted.