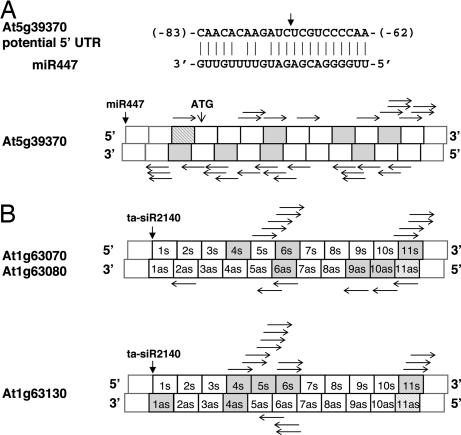
Fig. 3.
Positions of phased and nonphased small RNAs on candidate TAS genes. Small RNAs represented in the MPSS database and clustered in 21-nt increments for At5g39370 in rdr2 MPSS (A) and At1g63070/At1g63080/At1g63130 in Col-0 MPSS (B) data are indicated as gray boxes. Striped boxes correspond to the small RNAs whose start sites were used to determine the phased positions and to calculate the corresponding P value listed in Tables 2 and 3. The validated cleavage sites directed by ta-siR2140 from TAS2 are marked by vertical arrows for At1g63070/At1g63080/At1g63130. The predicted cleavage site by miR447 on At5g39370 and the position of its initial codon (ATG) are also marked. Labels in each box indicate the predicted ta-siRNAs derived from the 3′ cleavage product of the candidate TAS genes. Horizontal arrows mark all small RNAs (less than or equal to six hits) not in phase but represented in the MPSS databases. s, sense strand; as, antisense strand.