FIG. 2.
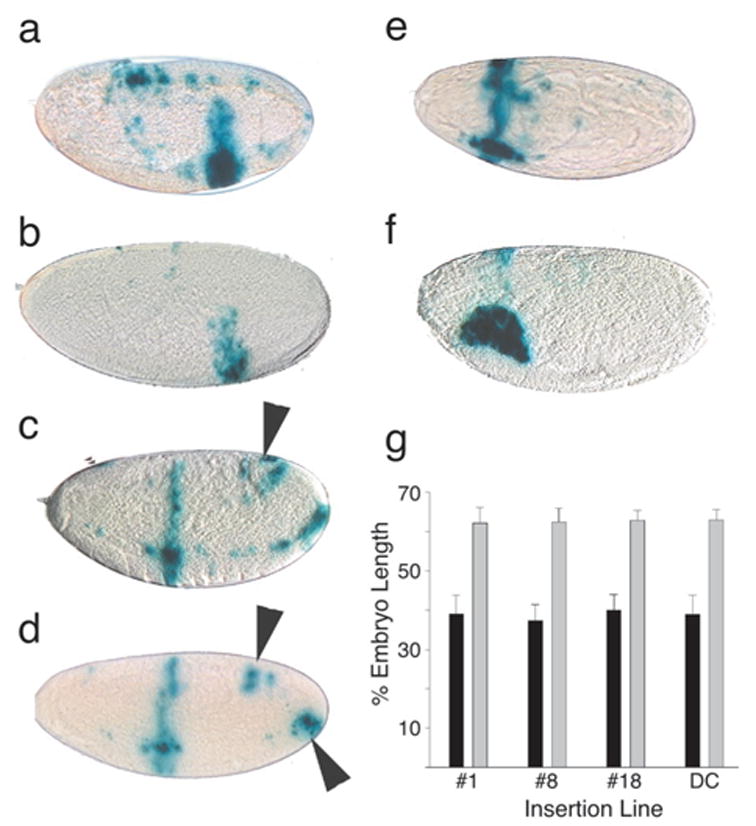
Expression patterns of Gal4 lines generated with the even-skipped stripe 3 enhancer. The stripe 3 enhancer was a 500 bp BamHI/SstI fragment derived from pPB-500st.3 and inserted into pGEM-3Zf(+) to acquire suitable restriction sites. The enhancer was then transferred as an XbaI/EcoR1 fragment to pPTGAL. Four transformant lines were recovered after injection (as described by Spradling, 1986) into early-stage embryos, three with insertions on the second chromosome (lines DC, 1 and 8) and one on the third (line 18). Shown are X-gal stained embryos of line 8 (a,c,e) and of line 18 (b,d,f) crossed to the second chromosome UAS-lacZ reporter strain (Brand and Perrimon, 1993). Eggs from 2-h collections were aged to give 4–6-h embryos (a,b), 8–10-h embryos (c,d), or 10–12-h embryos (e,f). g: Stripe position as a percentage of embryo length was determined from microscopic images with ImagePro software. Measurements were made from the anterior margin of the stripe to the anterior end of the embryo. The positions of anterior stripes (black bars, n = 15 embryos per line) are very consistent between insertion lines, as are the posterior stripes (gray bars, n = 16 embryos per line). Error bars indicate standard deviations.
