Figure 2.
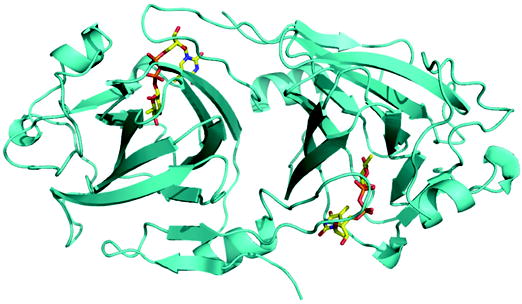
(a) Ribbon diagram of RmlC dimer from P. aeruginosa in complex with product (dTDP-6-deoxy-L-lyxo-4-hexulose). The product (an equatorially configured sugar nucleotide) is drawn in stick representation. The ligand atoms are shown in stick representation, with carbon yellow, nitrogen blue, oxygen red and phosphorus orange.
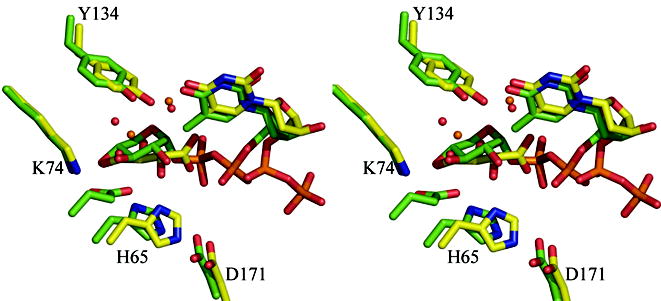
(b) Superposition of the dTDP-D-xylose complex with RmlC from P. aeruginosa and from S. suis 19. The β phosphate of dTDP-D-xylose in the P. aeruginosa is twisted out of the active site and a tartrate molecule occupies the active site in one monomer. In the S. suis structure the carbohydrate occupies the active site but is bound to E78 (shown but not labeled), a non-conserved residue on a loop not found in any other RmlC structure. The color scheme for P. aeruginosa complex is as Figure 2(a). For S. suis carbon atoms are colored green, other atoms are colored as P. aeruginosa. Water molecules in the P. aeruginosa complex are shown as orange spheres, in the S. suis they are shown as red spheres. Amino acid numbering is for the P. aeruginosa sequence.
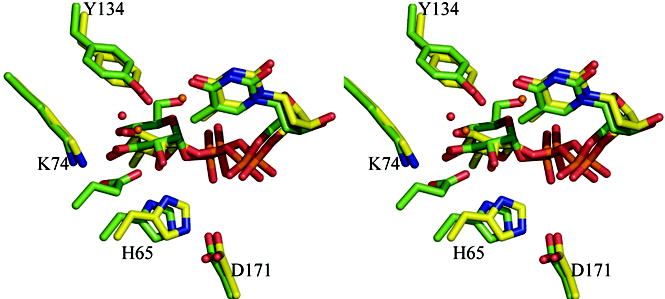
(c) Superposition of the dTDP-6-deoxy-L-lyxo-4-hexulose (product) complex with RmlC from P. aeruginosa and the dTDP-glucose complex from S. suis 19. Although the carbohydrate is bound at the active site in both complexes, the equatorial linkage between sugar and nucleotide in the P. aeruginosa complex means the sugar ring is positioned differently with respect to the key catalytic residues. The P. aeruginosa complex aligns better to the proposed chemical role of the active site residues. The color scheme is as Figure 2(b). The O6′ of dTDP-D-glucose from S. suis replaces one of the conserved structural water molecules and is bound by E78 (shown but not labeled). Amino acid numbering is for the P. aeruginosa sequence.
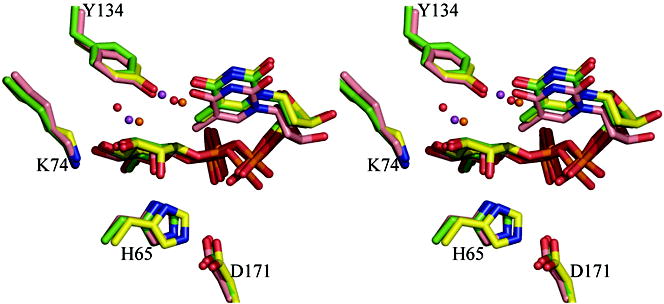
(d) Superposition of the dTDP-L-rhamnose complexes for three enzymes M. tuberculosis, S. suis and P. aeruginosa. The match between the structures is particularly striking especially given the different crystallization conditions and low sequence identity between the structures (25%). Two water molecules one close to C3′ and one close to C5′ are structurally conserved. The S. suis and P. aeruginosa structure are colored as Figure 2(c). The carbon atoms in M. tuberculosis RmlC complex are colored pink; other atoms are colored as P. aeruginosa. The two water molecules in the M. tuberculosis structure are shown as magenta colored spheres. Amino acid numbering is for the P. aeruginosa sequence.
