Fig. 8.
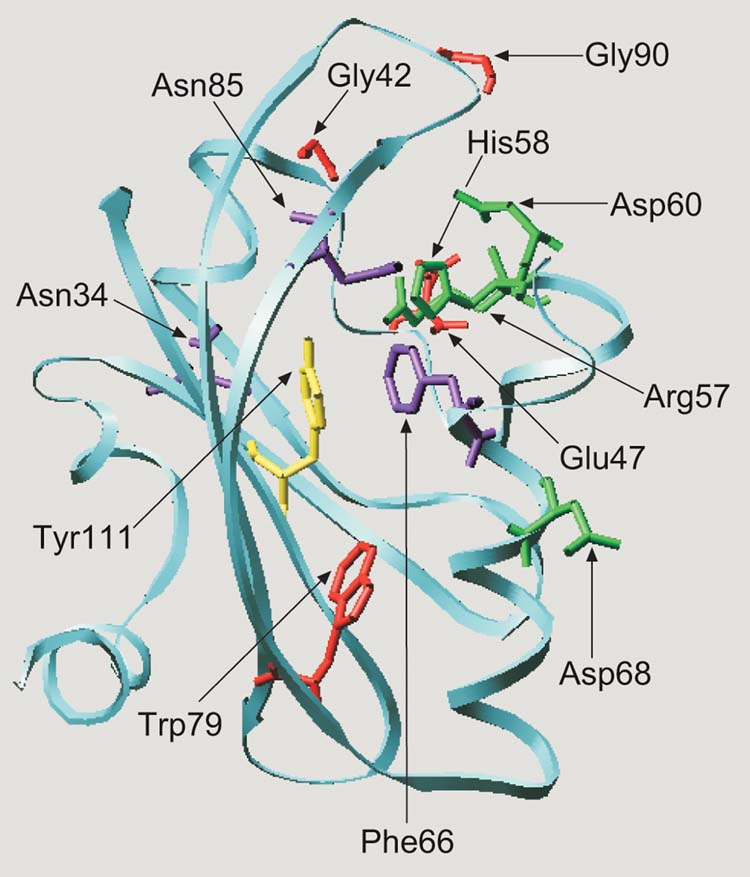
Hypothetical structural model for the E.coli Aer protein visualizing the predicted position of residues involved in aerotaxis. The Aer PAS domain (residues 1 to 119) was aligned with the PYP sequence (residues 1 to 125). The alignment was adjusted to minimize energy based on a sudo-Sippl field and submitted to the ProMod II program using the SWISS-PDB viewer. The figure was generated using the MOLSCRIPT program (Kraulis, 1991). Cysteine replacement of the residues shown as stick models produced a null aerotaxis phenotype (red), a loss of FAD binding and a null phenotype (green), inverted responses (yellow) and a CW signaling bias (purple).
