Figure 2.
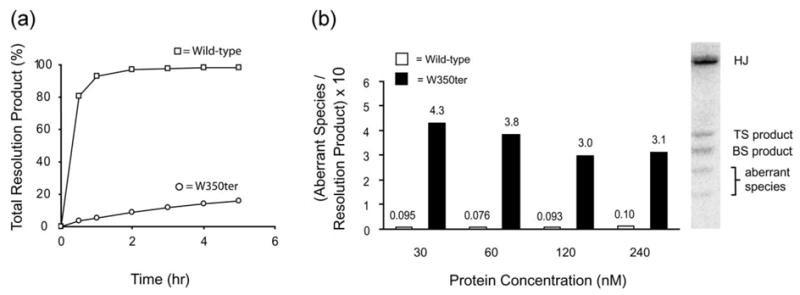
The C-terminal tail is required for efficient and accurate Holliday junction resolution. (a) Amount of total Holliday junction resolution product (top and bottom strand cleavages) by wild-type Int or W350ter. (b) Ratio of aberrant species to total Holliday junction resolution product at indicated protein concentrations of wild-type Int (open bars) and W350ter (filled bars). The aberrant species are likely single arm hairpins generated by uncoordinated Int cleavages, either during or after resolution.14,16 All Holliday junction resolution reactions ((a) and (b) and Figure 3(a)) were incubated at 20 °C and contain 120 nM integrase in assay volumes ranging from 10–100 μl with 50 mM Tris–HCl (pH 7.5), 50 mM NaCl, 0.5 mg/ml bovine serum albumin, 5 mM dithiothreitol, 10 nM 32P-labeled Holliday junction substrate, and 240 nM P′ 1,2 arm oligonucleotides. Reactions were quenched with a SDS gel loading solution and separated on 7% (w/v) polyacrylamide Tris–glycine (pH 8.0), 0.1% (w/v) SDS gels. Gels were subsequently dried and analyzed with the Fuji BAS-2500 phosphorimager system. Wild-type and mutant Int proteins were prepared as described.23 Int mutations were created via site-directed PCR mutagenesis with a Quikchange kit (Stratagene). Protein concentrations were determined by comparing the intensities of Coomasie-stained Int bands on an SDS-PAGE gel with those of a reference Int preparation quantified by amino acid composition (WM Keck Facility, Yale University). Preparation and sequences of synthetic Holliday junctions and P′ 1,2 oligonucleotides have been described.14
