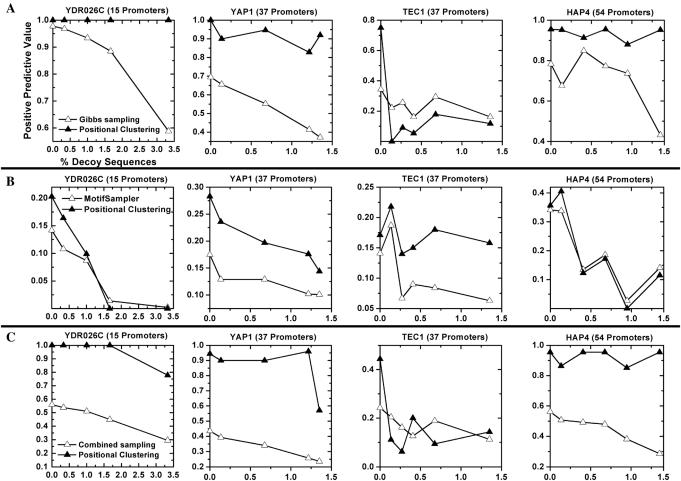
Figure 3.
Improvement and robustness of positional clustering on promoters bound to other yeast TFs. Sets of S.cerevisiae promoters bound by the TFs YAP1, TEC1, HAP4 and YDR026C were chosen according to ChIP-chip experiments (10). For each set, the initial promoters were analyzed using Gibbs sampling with positional clustering (solid triangles) and without (open triangles). Two Gibbs sampling approaches were applied to each dataset: a Gibbs sampler procedure similar to BioProspector (8) (row A), and MotifSampler (39) (row B). Row C shows the combination of both sampling procedures, along with positional clustering of the combined results. x-axis counts over addition of DSs. We evaluated the positive predictive value of each technique on each dataset, and found positional clustering generally improved the PPV through addition of 100% random DSs.