Figure 2.
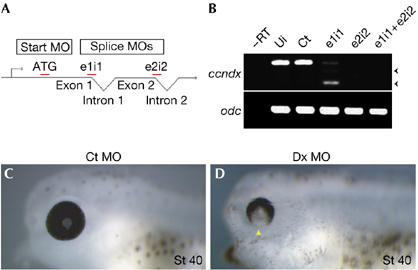
Phenotype of ccndx-deficient embryos. (A) Diagram depicting the target location of the different MOs. (B) Reverse transcription (RT)–PCRs of MO-injected embryos. MOs targeting splice junctions result in the shift of bands (truncated, arrowheads) or absence of transcripts relative to the controls. Ui, uninjected embryos; Ct, 5-mismatch e1i1 control MO. (C,D) Embryos injected with ccndx (Dx) MOs have defects in the formation of the ventral retina (D, arrowhead), relative to control MO-injected embryos (C). Ct MO, 5-mismatched e1i1 control MO; Dx MO, e1i1+e2i2 combined MOs; MO, morpholino antisense oligonucleotides; St 40, stage 40.
