FIGURE 4. Analysis of p62 suppression on in vitro differentiated osteoclasts treated with CLIC-5b oligonucleotides.
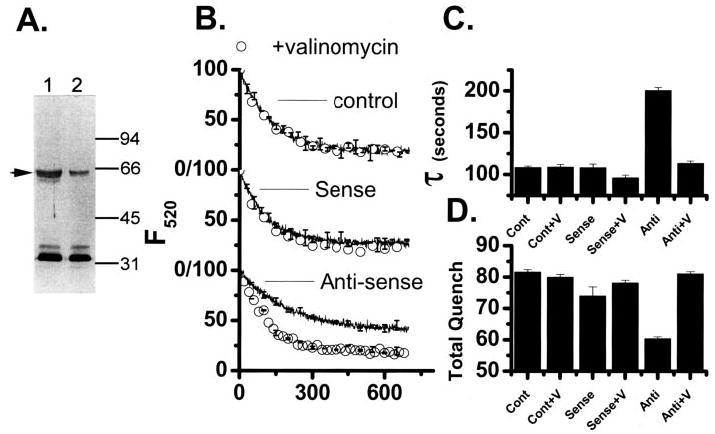
A, equal quantities of membrane protein from cells treated with sense (lane 1) or antisense (lane 2) CLIC-5b oligonucleotide were separated by SDS-PAGE and probed with antibody AP656 as previously described (15). Migration position of molecular mass standards are marked in kDa. For comparison the integrated intensities of the gels were normalized to those of the sense-treated cells in lane 1. The integrated density of the CLIC-5b protein band was decreased 71% by treatment with antisense oligonucleotide, while the bands between 32 and 34 kDa were unchanged. B, comparison of acidification by membrane vesicles prepared from cells treated with streptolysin-O in the absence of CLIC-5b oligonucleotide (top), cells treated with sense nucleotide (center), or cells treated with antisense nucleotide (bottom). Each figure compares acidification, (line) with acidification in the presence of 1 μM valinomycin (○). In addition to the data shown, acidification by each preparation was completely inhibited by bafilomycin A1 and displayed valinomycin reversed inhibition by substitution of sulfate for chloride as in Fig. 1. For each preparation the acidification-dependent quenching was verified to be reversible by the addition of NH4Cl (3, 15). The data were analyzed by curve fitting as described under “Materials and Methods” to determine the time constant of acidification (τ) and extent of acidification dependent quenching (total Quench). The antisense suppression was studied in three independent experiments using parallel control and sense treatments in every case. Each oligonucleotide treatment (control, sense, and antisense) was done in triplicate, and the cell membrane vesicles prepared and analyzed independently. The acidification curves shown are the average of the replicates for one representative experiment (n = 3) with the standard deviation presented for every 50th point. C, comparison of the time constants, τ, for vesicle preparations from cells treated as follows: control cells, Cont; control cells and added valinomycin, Cont+V; sense oligonucleotide-treated cells, Sense; sense oligonucleotide-treated cells and added valinomycin, Sense+V; antisense oligonucleotide-treated cells, Anti; and antisense oligonucleotide-treated cells and added valinomycin, Anti-V. D, total quench, for vesicle preparations that are labeled as in C. C and D summarize our total results from three independent experiments where each oligonucleotide treatment was done in triplicate. Each column is the average of the successful replicates from the three experiments, and the error bars are the S.D. for n = 3–9 but never less than 3.
