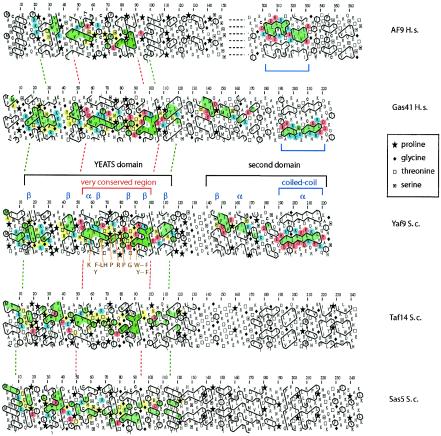
FIG. 2.
HCA indicates that Yaf9 is more similar to human (H.s.) Gas41 than to human AF9 or two other YEATS-domain proteins in S. cerevisiae (Taf14 and Sas5). HCA is a visual method that predicts protein secondary structure (9, 37). The linear one-dimensional sequence of the protein is written on an alpha-helix displayed along a cylinder. The cylinder is then cut parallel to its axis and unrolled in a bidimensional diagram. This diagram is duplicated in order to restore the full environment of each amino acid. The contours of clusters of hydrophobic residues are then drawn. This process has been automated by an online server (http://smi.snv.jussieu.fr/hca/hca-form.html). The alpha-helical net offers the best correspondence between the positions of hydrophobic clusters and regular secondary structures (alpha-helix and β strand). The HCA program uses the standard one-letter code for amino acids except for proline (regular secondary structure breaker), glycine (the least constrained amino acid), and serine and threonine (which can be accommodated in either hydrophilic or hydrophobic environments). Sequence features that were found in similar positions in Yaf9 and the other YEATS domain proteins were colored as follows: common hydrophobic clusters are in green, common nonhydrophobic amino acids are in yellow, positively charged amino” acids are in blue, and negatively charged amino acids are in red. “α” and “β” indicate the position of alpha-helical and β-sheet secondary structures for Yaf9 as predicted by the HCA analysis. Blue brackets show the positions of predicted coiled-coil sequences in the C-terminal regions of Yaf9, Gas41, and AF9. The global analysis of the sequences shows that these proteins are composed of two domains: a similar N-terminal region that corresponds to the YEATS domain and a C-terminal region that is more divergent. For Yaf9, Taf14, and AF9, a linker region could be clearly seen between these two domains. Examination of the HCA plots of human Gas41, AF9 and S. cerevisiae Yaf9, Taf14, and Sas5 sequences reveals the presence of similar hydrophobic motifs in the YEATS domain. The boundaries of this domain in the different proteins are shown with a dotted green line. Residues 55 to 100 of the Yaf9 sequence indicate an extremely conserved region shown within the dotted red lines. This central part of the first domain of the proteins was found to be characteristic of the YEATS domain family. The CLUSTALW (Fig. 1C) and HCA analyses show that the yeast Yaf9 and the human Gas41 YEATS domains are most similar to each other, whereas the yeast Taf14 and Sas5 YEATS domains are more similar to each other than either is to the remaining proteins. The second part of these proteins is more divergent and no similarity (>30%) in primary sequence could be found. Furthermore, the HCA profiles of Sas5, Taf14, and Yaf9 are different. On the other hand, similar hydrophobic motifs are found for the second domain of Yaf9 and Gas41. First, there are two hydrophobic clusters (green) separated by a conserved proline. Some amino acids, in particular charged residues, around this motif are well conserved. Second, a long alpha-helix predicted to form a coiled coil (blue brackets) is found at the C terminus of both Yaf9 and Gas41. We noted, however, that the charge distributions around these coiled coils are different, suggesting that their binding specificity may have diverged. Human AF9, both by its length (more than twice the length of the other YEATS proteins shown here) and by the profiling of its domains, seems the least similar of all of the YEATS proteins shown here.