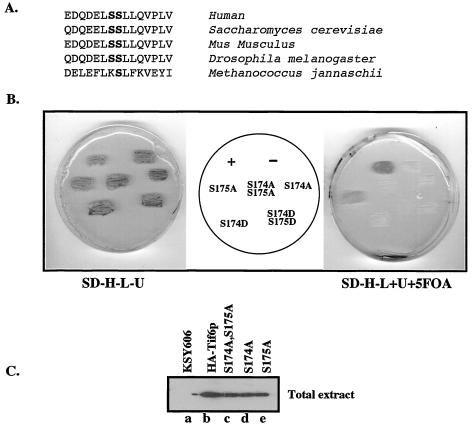
FIG. 4.
Effect of mutation of Tif6p at Ser-174 and Ser-175 on yeast cell growth and viability. (A) Conservation of the amino acid residues surrounding the serine CK I phosphorylation sites of eIF6 (Tif6p). Ser-174 and Ser-175 are in bold. (B) Haploid yeast strain KSY606, carrying an inactive chromosomal TIF6 gene and harboring the wild-type URA3-based TIF6 expression plasmid pRS316-TIF6 that expresses untagged Tif6p, was transformed with different LEU2-based TIF6 expression plasmids. These LEU2 plasmids expressed either HA-tagged wild-type Tif6p (+), HA-tagged mutant Tif6p (Ser-to-Ala or Ser-to-Asp mutation as shown), or no Tif6p ORF (−). Transformants were initially selected on SD−His−Leu−Ura plates (SD−H−L−U) and then replica plated onto (i) SD−His−Leu−Ura and (ii) SD−His−Leu plates with the addition of Ura and 5-FOA (SD−H−L+U+5FOA). The plates were then incubated at 30°C for 3 and 5 days, respectively. (C) Haploid yeast cells harboring both the URA3 plasmid pRS316-TIF6 and the different recombinant LEU2 expression plasmids expressing either the HA-tagged wild-type or HA-tagged mutant (S174A; S175A; and S174A, S175A) Tif6p proteins were recovered from the SD−His−Leu−Ura plates and grown to mid-logarithmic phase in SD medium. Cell lysates were then analyzed by Western blotting by using anti-HA antibodies. Lysates prepared from KSY606 cells that expressed untagged wild-type Tif6p were also analyzed as a control.