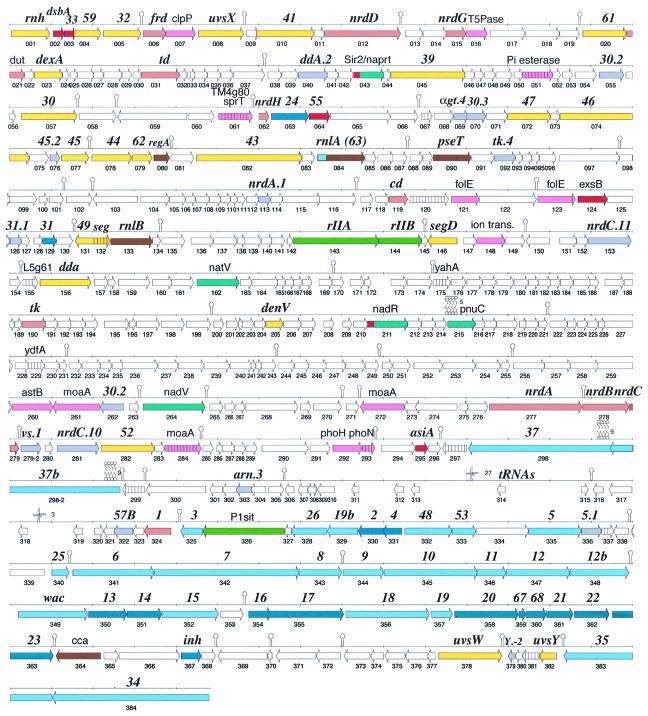
FIG. 1.
KVP40 CDS map. CDSs are labeled with the name of the closest homolog, and thus T4 nomenclature is primarily indicated (bold italics); nonitalics are primarily bacterial orthologs. Numbering below the CDS arrow is that used in the text, tables, and GenBank. Colors correspond to protein functional categories, as follows: orange, DNA metabolism; yellow, DNA replication; red, transcription; brown, translation; light blue, tail and tail fibers; dark blue, head; green, host interactions; turquoise, NAD salvage; pink, bacterial-type genes; gray, hypothetical genes in T4; hatched, conserved hypothetical; white, unique hypothetical. Stem-loops are predicted transcription terminators (Table 5) or other RNA structures; the clover leafs mark the positions of tRNAs, and the circle-squiggle symbols denote predicted transmembrane domains.