FIG. 1.
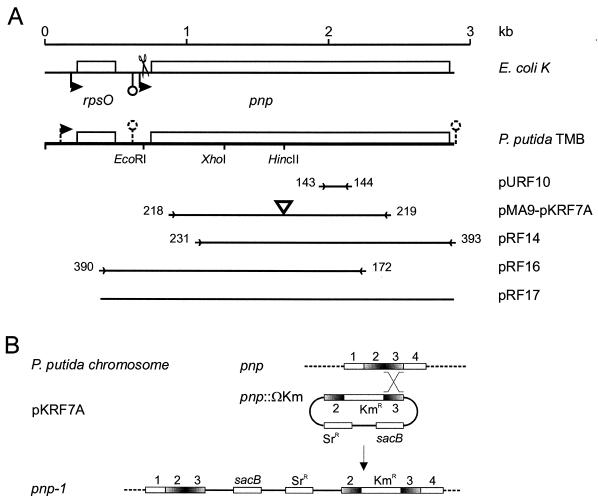
(A) Map of P. putida TMB rpsO-pnp region. The E. coli K-12 region is included at the top for comparison. Relevant restriction sites and cloned regions are shown; the corresponding plasmids (designations on the right) are described in Table 1. Promoters and intrinsic terminators are indicated by bent arrows and lollipops, respectively (dashed lines indicate that such features are putative). The terminator downstream of pnp is outside the sequence established in this work and was inferred from the complete genomic sequence of P. putida KT2442 (accession number NC_002947). Scissors indicate the RNase III cut site in the E. coli mRNA. Primers (Table 2) are indicated by arrowheads flanked by the oligonucleotide numbers and delimit the regions amplified by PCR and cloned. The triangle in pMA9 and pKRF7A indicates the Ω-Km cassette cloned into the HincII site. The fragment cloned in pURF10 was obtained by PCR of P. putida TMB genomic DNA with degenerate primers FG143 and FG144 (Table 2) designed with two amino acid motifs (PK/GRREIGHG and KAA/PVAGIAMG) conserved in the PNPases of E. coli (SwissProt accession number P05055), Photorhabdus luminescens (P41121), Haemophilus influenzae (P44584), B. subtilis (P50849), and Streptomyces antibioticus (Q53597). (B) Mutagenesis of P. putida pnp gene. An internal fragment of pnp (regions 2 and 3) interrupted by a kanamycin resistance cassette and cloned in the suicide plasmid pKNG101 (giving pKRF7A) recombined with homologous region 3 of the chromosomal gene, producing a merodiploid with two mutant pnp alleles (pnp-1 mutation in PPM101 and PPM103).