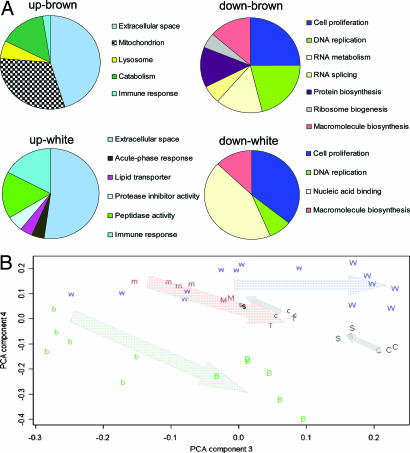
Fig. 3.
GO analysis of differentiating brown and white primary preadipocytes compared with C2C12 myoblasts and PCA. (A) Diagrammatic representation of the GO analysis carried out by using EASE. GO subgroups were collapsed into related groups to facilitate visualization of the data. For the complete breakdown of significantly enriched gene ontology groups, see SI Data Set 3. All GO groups demonstrated enhanced statistical representation (based on an EASE score and a Bonferroni-adjusted value of P < 0.05). (B) We used a PCA-based approach to compare the C2C12 differentiation data set, the SIRT1 differentiation data set (where SIRT1 overexpression prevents myoblast differentiation) with the differentiating pre-brown adipocytes and pre-white adipocytes. The data set includes day 4 undifferentiated brown (b) and white (w) preadipocytes and day 7/8 differentiating brown (B) and white (W) preadipocytes. C2C12 myoblasts are shown as m, and progressively differentiating myoblasts are plotted as M and T (http://pepr.cnmcresearch.org/). Sirt1-overexpressing cells (16) (s for 12-h differentiation and time control c, whereas S is 36-h differentiation with SIRT1 viral overexpression and time control C). After global normalization, we examined the top 100 genes that contributed to components 3 to 5 and used GO analysis to identify the putative SIRT1-regulated genes (see Methods). Components 3 and 4 are plotted. The arrows are used to illustrate the general direction of cell differentiation. It should be noted that SIRT1 (s and S) overexpression opposes the differentiation process.