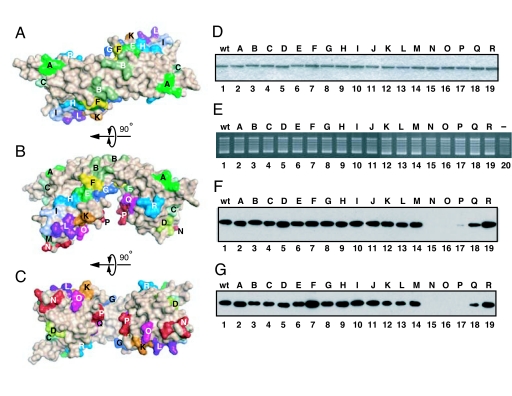
Fig. 4.
Mapping of SET/TAF-Iβ/INHAT activities. (A–C) Location of triple mutated residue sets as observed from the top view (A), side view (B), and bottom view (C) of SET/TAF-Iβ/INHAT. For detailed information, see SI Table 2. (D) SDS/PAGE of the SET/TAF-Iβ/INHAT WT and mutant proteins and staining with Coomassie brilliant blue. For full gel information, see SI Fig. 7. (E) Histone chaperone activities of the SET/TAF-Iβ/INHAT WT and mutant proteins. The estimated relative net activity of each mutant is as follows: WT, 100; A, 103; B, 94; C, 98; D, 87; E, 91; F, 97; G, 99; H, 97; I, 104; J, 98; K, 98; L, 93; M, 88; N, 38; O, 36; P, 67; Q, 64; R, 95 (see Materials and Methods). (F and G) Histone-binding (F) and dsDNA-binding (G) activities of the SET/TAF-Iβ/INHAT WT and mutant proteins. The bound proteins were detected by immunoblot analysis using anti-His antibodies. Mutants N, O, and P demonstrated impaired binding activities. The estimated relative histone binding activity of each mutant is as follows: WT, 100; A, 98; B, 101; C, 91; D, 127; E, 98; F, 108; G, 103; H, 101; I, 108; J, 106; K, 96; L, 81; M, 100; N, 0; O, 0; P, 5; Q, 70; R, 122. The estimated relative DNA binding is as follows: WT, 100; A, 71; B, 89; C, 114; D, 121; E, 123; F, 117; G, 124; H, 138; I, 88; J, 77; K, 79; L, 73; M, 64; N, 5; O, 7; P, 0; Q, 29; R, 59 (see Materials and Methods).