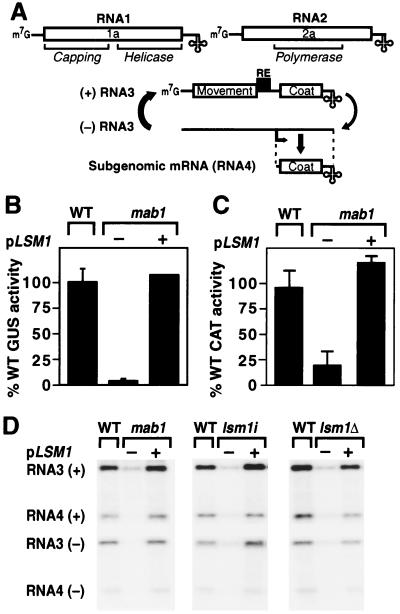
Figure 1.
Identification of LSM1. (A) Schematic of the BMV genome, RNA3 replication, and subgenomic mRNA synthesis, showing ORFs (boxed), noncoding regions (single lines), 5′ caps (m7G), intergenic replication enhancer (RE), and tRNA-like 3′ ends (cloverleaf). (B) BMV-directed β-glucuronidase (GUS) expression in 1a- and 2a-expressing wild-type (WT) and mab1 yeast containing a chromosomally integrated B3GUS expression cassette. Total protein was extracted, and GUS activity per milligram of total protein was measured. Averages and SEM from three experiments are shown, except for mab1 + pLSM1, which is the average of two experiments. pLSM1 is a centromeric yeast plasmid containing a 1.5-kilobase (kb) DNA fragment bearing WT LSM1. (C) BMV-directed chloramphenicol acetyl transferase (CAT) expression in mab1 and WT yeast expressing 1a and 2a and electroporated with B3CAT in vitro transcripts. Yeast was incubated in glucose medium and assayed for CAT activity 21 h after electroporation. Averages and SEM from three experiments are shown. (D) Northern blot analysis of positive- and negative-strand BMV RNA3 and subgenomic RNA4 accumulation in WT yeast, the original mab1 mutant strain, the lsm1i isogenic strain, and the lsm1Δ knockout strain expressing 1a, 2a, and WT RNA3. Equal amounts of total yeast RNA were loaded in each lane. The negative-strand PhosphorImager image was printed at higher sensitivity to approximate a 10-fold longer exposure than the positive-strand blot.