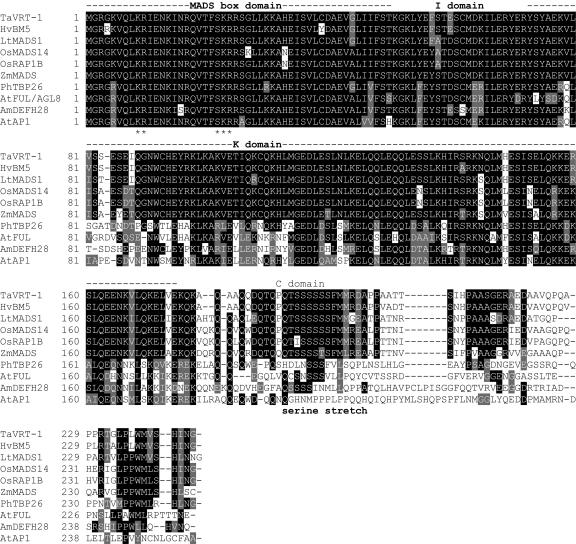
Figure 1.
Structure and sequence alignment of TaVRT-1 with other MADS-box members of the API/SQUA family. Barley (Hordeum vulgare) HvBM5 (Schmitz et al., 2000), Lolium temulentum LtMADS1 (Gocal et al., 2001), indica rice OsMADS14 (Monn et al., 1999), japonica rice OsRAP1B (Kyozuka et al., 2000), maize (Zea mays) ZmMADS from EST BE511439, petunia (Petunia hybrida) PhTBP26 (Immink et al., 1999), Arabidopsis AtFUL/AGL8 (Mandel and Yanofsky, 1995), Snapdragon (Antirrhinum majus) AmDEFH28 (Müller et al., 2001), and Arabidopsis AtAP1 (Mandel et al., 1992). Identical and similar amino acids are shaded in black and gray, respectively. MADS-box domain, DNA-binding domain; I, intervening region; K, keratin-like domain; C, C-terminal region. *, Residues identified as being part of a nuclear targeting signal by PSORT (Nakai and Kanehisa, 1992; http://psort.nibb.ac.jp). Ser stretch, NetPhos 2.0 (Blom et al., 1999) predicts phosphorylation sites in this region (http://www.cbs.dtu.dk/services/NetPhos/).