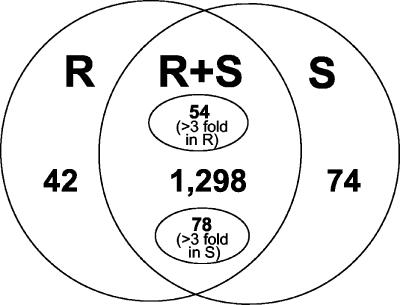
Figure 1.
Identification of differentially expressed genes by in silico EST subtraction. ESTs derived from the R11-12 (R) and the R11-13 (S) libraries were assembled into contigs. Within each contig, the number of ESTs derived from the R or S libraries was then calculated. Those contigs (with at least three ESTs) with ESTs from only the R library are shown on the left, and those with ESTs from only the S library are shown on the right. Contigs that contained mixtures of ESTs from both libraries are shown in the middle. Fifty-four of these contigs contained 3-fold or more ESTs from the R than S library, whereas 78 contigs contained 3-fold or more ESTs from the S than the R library. The remainder of the contigs (1,298) contained mixtures of ESTs from both libraries but in less than 3-fold ratios.