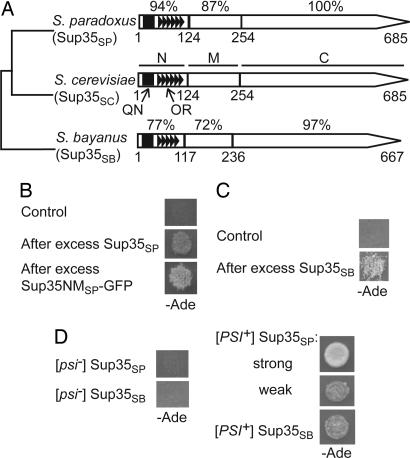
Fig. 1.
S. paradoxus and S. bayanus Sup35 proteins retain prion-forming abilities despite divergence of PrD sequences. (A) Structural and functional organization of the S. sensu stricto Sup35 proteins. N, M, C, QN, and OR refer to the Sup35N, Sup35M, Sup35C, QN-rich stretch, and ORs, respectively. Numbers correspond to amino acid positions; the percentage of amino acid identity to S. cerevisiae is shown for each region of S. paradoxus and S. bayanus proteins individually. For sequence alignment of the Sup35N regions, see SI Fig. 5. Data are from refs. 28–30 and the Yeast Genome Database (www.yeastgenome.org), confirmed by sequencing of our PCR-amplified clones. (B and C) Transient overproduction of S. paradoxus Sup35 protein (Sup35SP) or the Sup35NMSP-GFP fusion protein (B), or transient overproduction of S. bayanus Sup35 protein (Sup35SB) (C) induces prion formation in the [psi− PIN+] S. cerevisiae strains bearing the SUP35SP (B) or SUP35SB (C) gene instead of SUP35SC. Empty plasmids pmCUP-GFP (B) or pRS316GAL (C) were used as controls. Prion formation was detected by growth on −Ade medium after induction on PCUP1-SUP35SP or PCUP1-SUP35NMSP-GFP constructs on the medium with 100 μM CuSO4 (B) or PGAL-SUP35SB construct on Gal medium (C). (D) Sup35SP generates both strong and weak prion variants, whereas Sup35SB generates only weak prion variants in S. cerevisiae, as judged from the efficiency of ade1–14 suppression reflected by growth on −Ade. Note that both strong and weak variants of the Sup35SP prion show low mitotic stability (see SI Table 2). Plates were photographed after 5 (B), 8 (C), and 7 (D) days of incubation.