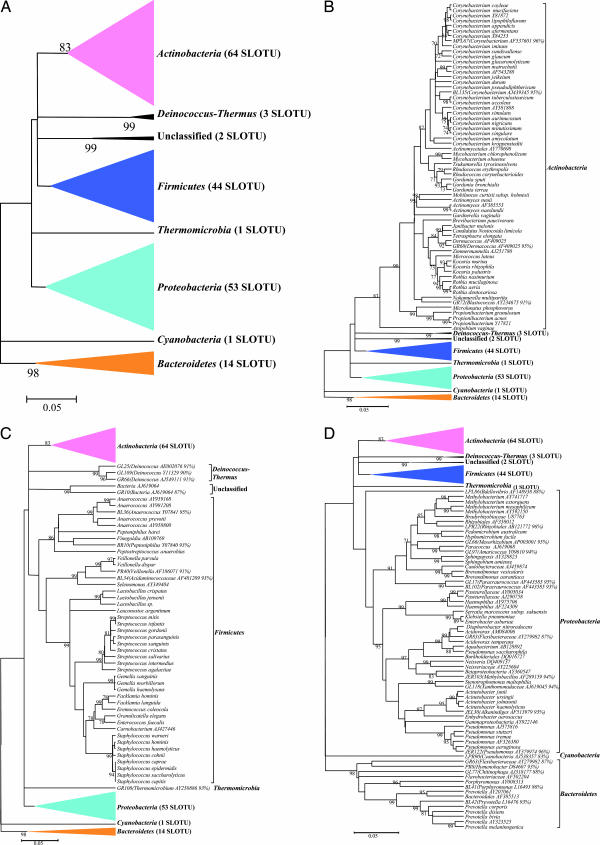
Fig. 1.
Phylogenetic analysis of bacterial 16S rDNA detected in normal skin from six subjects. (A) From 1,221 clones, sequences representing eight bacterial phyla and 182 SLOTUs were observed. Alignments were done with Greengenes, and misalignments were manually curated in ARB (48), evolutionary distances were calculated with the Jukes–Cantor algorithm, and phylogenetic trees were determined by the Neighbor-Joining method; with 1,000 trees generated, bootstrap confidence levels are shown at tree nodes for values ≥70%. (B) Phylogenetic tree of the 628 clones within the phylum Actinobacteria. SLOTU designations are located at the termination of each branch. The 16S rDNA clones and cultivated non-type strains represent potential bacterial species; these clones are represented by the nearest species, followed by the GenBank accession number of the best-matched sequence. Unknowns are represented by the serial number of the clone used in this study, followed by the closest match, and percent sequence identity. (C) Phylogenetic tree of the 345 clones representing the phyla Firmicutes, Deinococcus-Thermus, Thermomicrobia, and unclassified organisms. Designations are as described for B. The clones representing Deinococcus-Thermus, Thermomicrobia, and unclassified were from the same subject. (D) Phylogenetic tree of the 248 clones representing the phyla Proteobacteria, Bacteroidetes, and Cyanobacteria. Designations are as described for B.