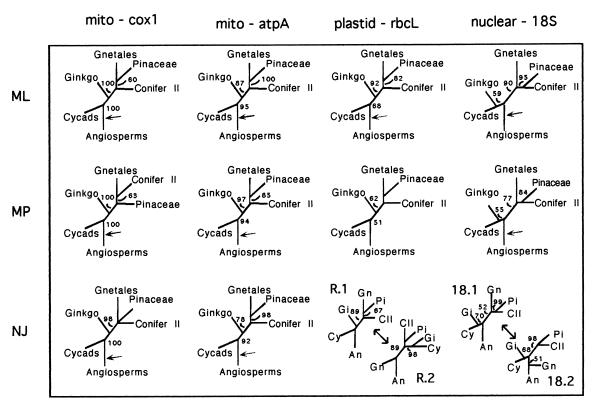
Figure 2.
Alternative phylogenetic analyses of four genes by using ML, MP, and NJ. Analyses are unrooted, with arrows indicating position of root in additional rooted analyses (see text for details). BS above 50% for each analysis are given on internal branches. Taxa and tree characteristics are as Fig. 1, except long-branch taxa (Ephedra, Podocarpus, Phyllocladus) are excluded for cox1. Additional tree characteristics for each analysis: for cox1: 23 taxa, ML: -ln likelihood = 4827.7315 (ti/tv = 1.464, inv sites = 0.335, gamma = 0.758); MP: 1 tree at 502 steps, CI = 0.707, RI = 0.850; NJ: min. evol. score = 0.5217; for atpA: 16 taxa, 1,283 and 228 bases; ML: -ln likelihood = 5,118.0457 (ti/tv = 1.783, inv sites = 0.542, gamma = 3.911); MP = 1 tree at 639 steps; CI = 0.655, RI = 0.767; NJ: min. evol. score = 0.8169; for rbcL: 26 taxa, MP: 3 trees at 1,329 steps; CI = 0.438, RI = 0.637; NJ: min. evol. score = see text; for 18S: 27 taxa, MP: 2 trees at 780 steps, CI = 0.499, RI = 0.717; NJ: min. evol. = see text.