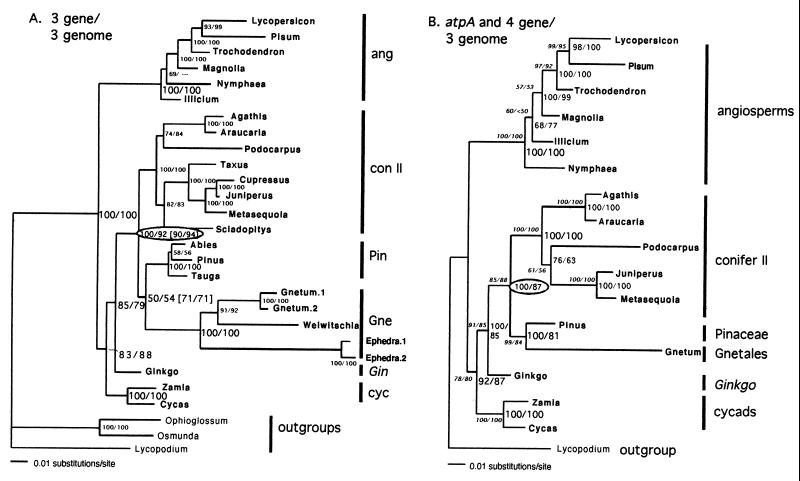
Figure 3.
Rooted seed-plant phylogenies by using three-genome combined data from (A) cox1, rbcL, and 18S rDNA, and (B) including atpA. Identical topologies were obtained by using ML, MP (except arrangement of angiosperms, where Nymphaea is basal in three-gene MP), and NJ analyses, rooted or unrooted, with or without long-branch Ephedra. (A) ML/MP bootstraps are shown. Twenty-eight taxa; 4,367 and 1,025 bases; ML: –ln likelihood = 24,645.044 (ti/tv = 2.137, inv sites = 0.427, gamma = 0.685); MP: 3 trees at 3,583 steps (CI = 0.49; RI = 0.67). ML/MP bootstraps also given for two key nodes (in brackets) after deletion of long-branch Ephedra. (B) ML/MP bootstraps shown for atpA alone (Left) and all four genes (Right). For atpA (17 taxa, 1,283 and 244 bases): ML: -ln likelihood = 5,678.420 (ti/tv = 1.774, inv sites = 0.431, gamma = 1.836; MP: 1 tree at 777 steps (CI = 0.621; RI = 0.723). For all 4 genes (5,650 and 920 bases): ML: -ln likelihood = 23,323.1563 (ti/tv = 2.224, inv sites = 0.508, gamma = 0.938); MP: 1 tree at 3,044 steps, CI = 0.549, RI = 0.641.