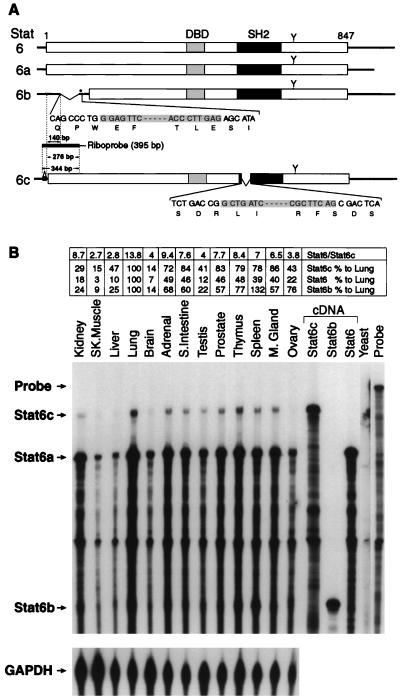
Figure 1.
(A) Schematic diagram of Stat6, Stat6a, Stat6b, and Stat6c cDNAs. The Stat6-encoded cDNA (35) is illustrated as an open box. The putative DNA binding domain (DBD), SH2 domain, and a known tyrosine residue phosphorylated upon IL-4 treatment (Y) are indicated. Bases deleted in Stat6b and Stat6c are inclusively shaded. In Stat6b, an in-frame stop codon is designated by an asterisk. The riboprobe spanning the Stat6c insertion/Stat6b deletion used in the RNase protection assay and designed to protect hybrid products of 276, 140, and 344 bp for Stat6, Stat6b, and Stat6c, respectively, as described in Materials and Methods is depicted. (B) RNase protection assay of Stat6 variant RNAs isolated from human tissues. Total RNA prepared from several human tissues was used in the RNase protection assay. The protected products were analyzed on 6% acrylamide–urea gels, and the dried gel was exposed to x-ray film for 24 h with intensifying screens. The GAPDH antisense probe (97 bp) was run as an internal standard for each tissue and exposed for 3 h. Stat6, Stat6b, and Stat6c cDNAs were also subject to RNase protection and shown as controls. Quantitative comparison of Stat6 transcripts was performed by excision of each fragment, liquid scintillation counting, and normalization to the internal GAPDH antisense probe. The relative amount of each Stat6 variant transcript was quantitated relative to lung tissue (100%) for the same variant. Additionally, the relative ratio of Stat6/Stat6c transcript is presented.