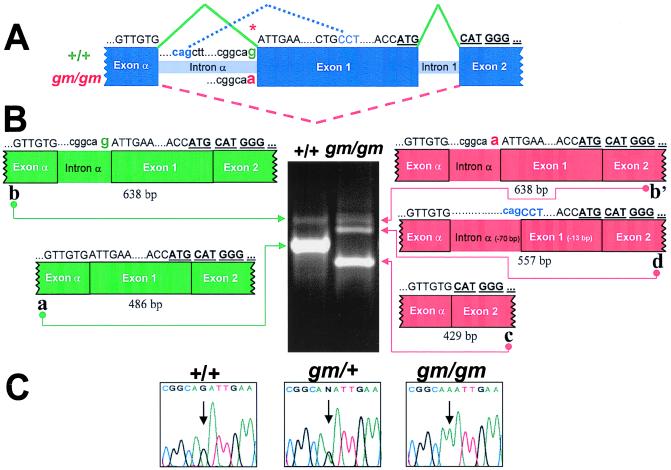
Figure 3.
A splice acceptor mutation in Rabggta causes abnormal splicing. (A) Genomic structure of the 5′ end of mouse Rabggta. Exon α encodes 5′ untranslated region. The initiation methionine (bold and underlined ATG) occurs at the 3′ end of exon 1. Rabggta nucleotide sequence above is C57BL/6J-+/+ and below is C57BL/6J-gm/gm. The splice acceptor mutation is indicated by *. Rabggta exons connected by solid green lines represent normal splicing. Dashed red lines indicate exon 1 skipping in gm/gm RNA, and dotted blue lines indicate cryptic splice sites used by gm/gm. (B) RT-PCR analysis of RNA from wild-type (+/+) and mutant (gm/gm) bone marrow. The Rabggta oligonucleotides used amplified the region between exon α and exon 2. Each band was excised and sequenced: Normal bone marrow (+/+) shows the expected product of 486 bp (band a) (GenBank accession no. AF127656), and a product (638 bp) retaining intron α (band b) (GenBank accession no. AF127658). gm/gm bone marrow shows three bands: Band c (429 bp) (GenBank accession no. AF127657) represents exon 1 skipping; band b′ (638 bp) (GenBank accession no. AF127659) retains intron α; band d (557 bp) (GenBank accession no. AF127662) represents utilization of cryptic splice sites in intron α and exon 1. (C) Sequence analysis of Rabggta genomic DNA. Arrows indicate the splice acceptor mutation: +/+ = G, gm/gm = A, gm/+ exhibits both nucleotides (G and A).