Figure 2.
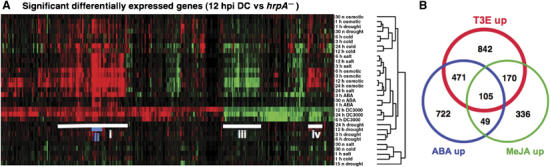
Hierarchal clustering of T3E-responsive transcripts shows a strong overlap with abiotic stresses. (A) The 880 probesets identified previously as significantly differentially expressed in response to T3Es at 12 hpi were clustered using GeneChip expression data from the AtGenExpress consortium. Experiments reporting the effects of cold, drought, salt stress, osmotic stress (mannitol) and ABA application were included as well as additional time points reporting T3E activity. All data sets were normalised and interpreted using the GCRMA function of the Bioconductor microarray analysis package (http://www.bioconductor.org/). Hierarchical clustering was applied using an uncentred correlation and complete linkage clustering. Genes induced relative to their control are coloured red, those suppressed are coloured green, whereas genes unchanged in their expression levels are coloured black. Cluster i, genes sharing strong similarity to regulatory networks induced by abiotic stresses, in particular, osmotic stress. Cluster ii, all the Clade A PP2Cs originally identified in Table I are highlighted in blue. Cluster iii, genes suppressed by both T3E and abiotic stresses. Cluster iv, genes suppressed by T3E but induced by abiotic stresses. (B) Venn diagram showing the commonality between transcripts differentially induced by T3Es and those strongly induced by ABA or MeJA application. SAM (Tusher et al, 2001) was used to identify genes induced by DC3000 relative to DC3000hrpA− or DC3000hrcC at 6, 12 and 24 hpi from two independent but similar experiments, with a minimum fold change of 2 and a false discovery rate of less than 5%. ABA- and MeJA-induced genes were identified solely on fold change taking the average of two replicates.
