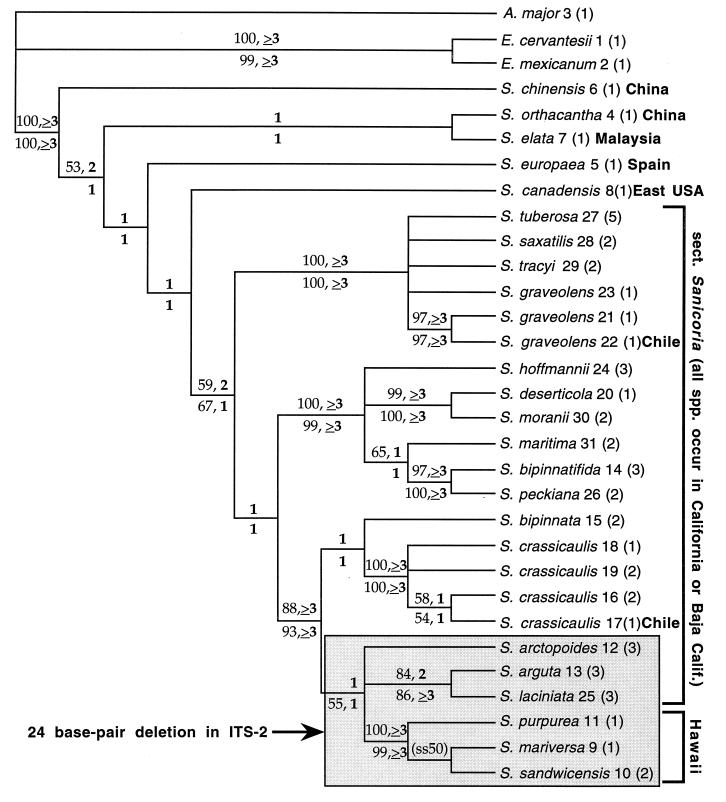
Figure 1.
Semistrict consensus of 252 minimum-length Fitch parsimony trees of nuclear ribosomal DNA ITS sequences in Sanicula and outgroups, without recoding of insertions/deletions (indels) as additional characters (consistency index = 0.74; retention index = 0.79). The same island of 252 trees was found in each of the 20 heuristic analyses, with random addition sequences of the taxa. Numbers immediately after species names refer to numerically designated sequences in Table 1. Numbers in parentheses indicate the number of populations sampled. Numbers above tree branches are bootstrap (standard type) and decay (boldface type) values from analyses of the data matrix (Table 1) with recoded indel characters excluded. Numbers below tree branches are bootstrap (standard type) and decay (boldface type) values from analyses of the data matrix (Table 1) including the recoded indel characters. Only bootstrap values above 50% are shown. A., Astrantia; E., Eryngium; S., Sanicula. (ss50) indicates the only branch not resolved in the strict consensus of minimum-length trees; the branch was resolved in 50% of the minimum-length trees and in the semistrict consensus tree.